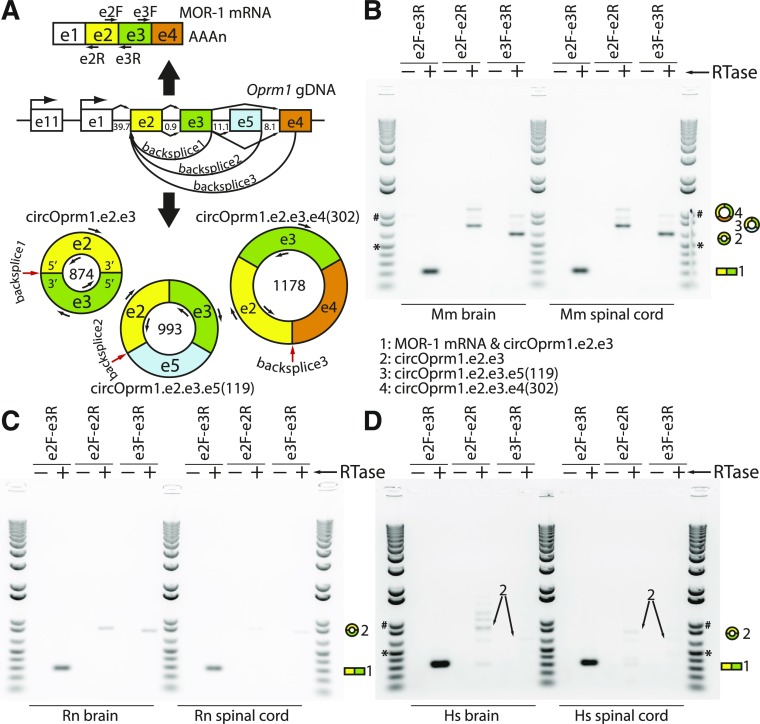
Fig. 1.
Mouse, rat, and human Oprm1/OPRM1 divergent RT-PCRs of circular RNAs. (A) A map of the mouse μ-opioid receptor gene (Oprm1) is shown with select exons in their relative locations (e.g., e1 designates exon 1). The e1 and exon 11 (e11) associated promoters are marked with bent arrows, and intron sizes are labeled in kilobases. The μ-opioid receptor (MOR-1) protein is encoded by the dominant linear mRNA containing e1-e2-e3-e4, shown above the genomic DNA (gDNA) with a polyA tail and locations of forward (F) and reverse (R) primers. The Oprm1 locus also generates multiple circRNA isoforms (shown below the gDNA map, with primer sites; circRNA sizes in nucleotides shown inside circles; red arrows point to backsplice junctions). (B) Divergent RT-PCR products from mouse [Mus musculus (Mm)] brain and spinal cord total RNA were Sanger sequenced and backsplice junctions confirmed (also see Supplemental Figs. 1 and 2). In brain, three variants were sequence confirmed, while only circOprm1.e2.e3 was confirmed in spinal cord. (C) Rat [Rattus norvegicus (Rn)] and (D) human [Homo sapiens (Hs)] divergent RT-PCRs were designed similarly to the mouse. Conventional RT-PCR with convergent primers (e2F-e3R) amplify linear mRNA as well as circRNAs, while divergent primers (e2F-e2R, e3F-e3R) only amplify circRNAs. In all cases, 5 μg total RNA was treated with reverse transcriptase [(RTase), arrow] with 250 ng random hexamers in 20 μl reactions (+), or RTase omitted (−). PCR was done with 1 ng/μl of cDNA input to 35 ampflication cycles. Ten microliters of PCR reaction were loaded against 1 μl of Invitrogen 1 kb Plus ladder; 500 bp (*) and 1 kb (#) are marked as size reference.