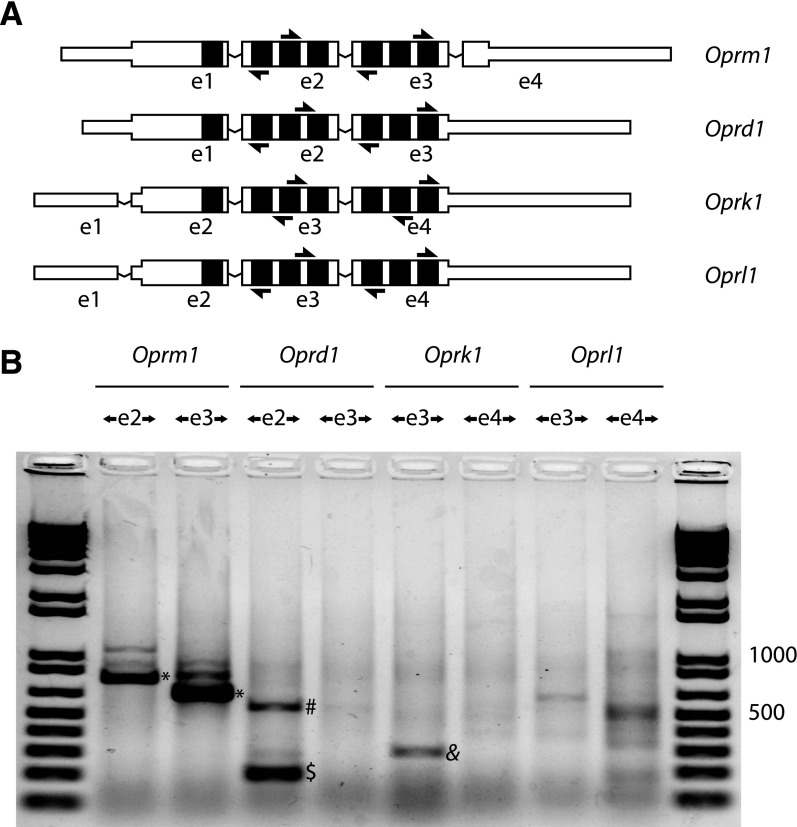
Fig. 3.
Mouse non-μ-opioid receptor divergent RT-PCRs. (A) A schematic of mouse μ (Oprm1), δ (Oprd1), κ (Oprk1), and nociceptin (Oprl1) receptor mRNAs for the dominant splice isoforms is shown, with exons labeled below (exon 1 is designated as e1). The mRNAs are organized by alignment of homologous exons, with transmembrane span coding regions colored black. Divergent RT-PCR primers for each target sequence are shown as arrows. Exons are not drawn to scale; narrow portions of exons represent untranslated regions. (B) Divergent RT-PCR was performed on mouse whole brain total RNA with PCR primer pairs directed outwardly from exons as labeled. For each gene, two primer pairs were used, targeting two separate exons. The Oprm1 locus generates several circRNAs, of which the e2.e3 circRNA appears dominant (*). Oprd1 e2 generates a single exon circRNA (dollar), and this template supported detection of a dimeric RT-PCR product consistent with processive reverse transcription of a single exon circRNA (#). Oprk1 also generates a single exon e3 circRNA (&). These divergent RT-PCRs were confirmed to derive from circRNAs by examination of backsplice sequences from direct Sanger sequencing, or after TOPO cloning, to confirm the existence of a backsplice junction and adherence to canonical GT-AG splice junctions. Five micrograms of total RNA were treated with reverse transcriptase using 250 ng random hexamers in 20 μl reactions, and 35 cycles of PCR amplification were done with 25 ng/μl input cDNA. Five microliters of PCR reaction were loaded against 1 μl of Invitrogen 1 kb Plus ladder; 500 and 1000 bp are labeled as size reference.