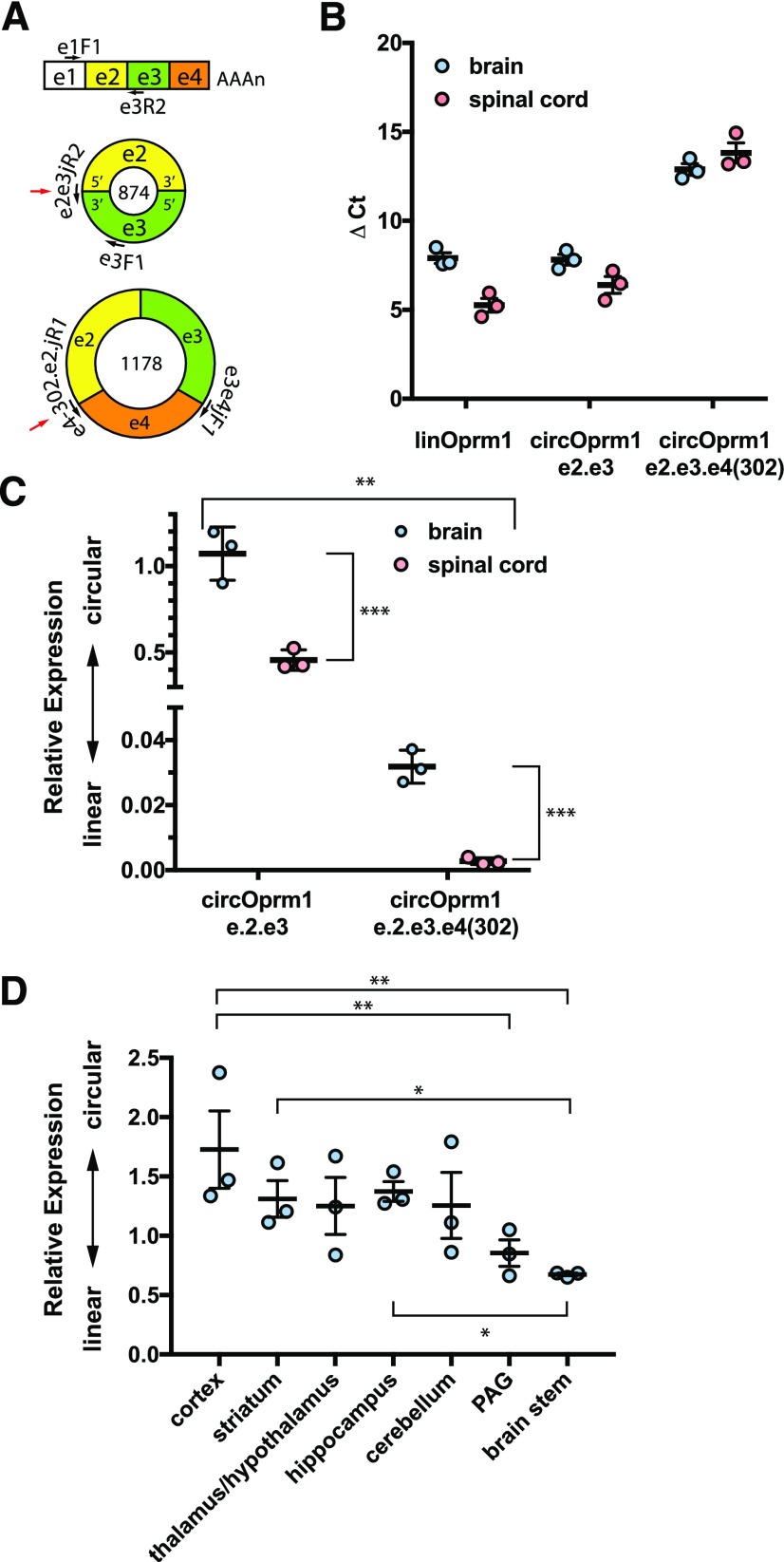
Fig. 4.
Anatomic survey of expression of mouse Oprm1 linear vs. circular RNAs. (A) The target Oprm1 RNAs are shown with exons and primers not drawn to scale. The linear Oprm1 isoforms are detected with primers spanning exon 1 (e1F1) to exon 3 (e3R2). The circOprm1.e2.e3 is detected with a backsplice junction spanning primer (e2e3juncR2) paired with an exon 3 forward primer (e3F1). The circOprm1.e2.e3.e4(302) is selectively amplified with two junction spanning primers. The backsplice junctions for circRNAs are shown with a red arrow. (B) ΔCt values for Oprm1 isoforms in opioid naive mouse whole brain and spinal cord are shown for linear, circOprm1.e2.e3, and circOprm1.e2.e3.e4(302). (C) Relative abundance values for circular isoforms normalized to the linear isoforms, using data shown in (B), are plotted as ΔΔCt values and analyzed by two-way ANOVA (significant variation was attributable to anatomic region, ***P < 0.001; RNA target, **P < 0.01; and interaction, **P < 0.01, see Materials and Methods). (D) Select brain regions were dissected from naive CD-1 male mice, and qPCRs were performed to evaluate relative expression of linear and e2.e3 circRNA levels (overall P < 0.05 by ordinary one-way ANOVA (F6,14 = 2.953; *P = 0.045), followed by post-hoc multiple comparisons analysis with Tukey’s correction significant for cortex vs. brain stem (*P < 0.05).