Abstract
Objective
Pancreatic β cell failure plays a central role in the development of type 2 diabetes (T2D). While the transcription factors shaping the β cell gene expression program have received much attention, the post-transcriptional controls that are activated in β cells during stress are largely unknown. We recently identified JUND as a pro-oxidant transcription factor that is post-transcriptionally upregulated in β cells during metabolic stress. Here we seek to uncover the mechanisms underlying this maladaptive response to metabolic stress.
Methods
RNA-protein and protein-protein interactions were measured using RNA immunoprecipitation and co-immunoprecipitation, respectively, in Min6 cells and mouse islets. Phos-tag analyses were used to assess hnRNPK phosphorylation in primary mouse and human islets and Min6 cells. Translating ribosome affinity purification (TRAP) followed by RT-qPCR was used to identify changes in the ribosome occupancy of mRNAs in Min6 cells. Gene depletion studies used lentiviral delivery of CRISPR-Cas9 to Min6 cells. Apoptosis was measured in primary islets using a cell-permeable dye with a fluorescence readout of activated cleaved caspase-3 and-7.
Results
A de novo motif analysis was performed on a subset of genes previously found to be regulated at the level of ribosome binding during PDX1-deficiency, which identified a poly-cytosine (polyC) motif in the 3′UTR of the transcript encoding JUND. The polyC-binding protein hnRNPK bound to the mRNA encoding JUND, leading us to hypothesize that hnRNPK regulates JUND expression during glucolipotoxicity. Indeed, loss of hnRNPK blocked the post-transcriptional upregulation of JUND during metabolic stress. hnRNPK was phosphorylated in mouse and human islets during glucolipotoxicity and in islets of diabetic db/db mice. The MEK/ERK signaling pathway was both necessary and sufficient for the phosphorylation of hnRNPK, upregulation of JUND levels, and induction of pro-oxidant and pro-inflammatory genes. Further, we identified the RNA helicase DDX3X as a new binding partner for hnRNPK that is required for efficient translation of JUND mRNA. Loss of hnRNPK reduced DDX3X binding to translation machinery, suggesting that these factors cooperate to regulate translation in β cells.
Conclusions
Our results identify a novel ERK/hnRNPK/DDX3X pathway that influences β cell survival and is activated under conditions associated with T2D.
Keywords: RNA-Binding proteins, β cell, Stress, Translational regulation, Signaling
Abbreviations: TRAP, Translating Ribosome Affinity Purification; GLT, Glucolipotoxicity
Highlights
-
•
A polyC motif is identified in the 3′UTRs of post-transcriptionally regulated genes.
-
•
hnRNPK mediates the post-transcriptional induction of JUND during metabolic stress.
-
•
hnRNPK is phosphorylated in β cells during metabolic stress.
-
•
MEK/ERK signaling is necessary and sufficient to activate hnRNPK/JUND.
-
•
DDX3X interacts with hnRNPK and regulates translation of JUND mRNA.
1. Introduction
The development of type 2 diabetes (T2D) in individuals with insulin resistance depends on the dysfunction and eventual loss of pancreatic β cells. This reduction in β cell number is caused by chronic exposure to conditions associated with T2D, including elevated serum levels of glucose and free fatty acids [1]. These changes in the extracellular environment have broad impacts on β cells, including alterations of gene expression networks. Indeed, it has been suggested that chronic stress not only can lead to β cell apoptosis but also can cause a loss of cell identity due to disruption of the β cell gene expression program [2], [3]. Therefore, identifying factors that shape gene regulatory networks in β cells during stress is central to our understanding of T2D pathogenesis.
An important class of factors that regulate gene expression programs is RNA binding proteins (RBPs). RBPs recognize specific sequence elements and/or RNA secondary structures typically located in the untranslated regions (UTRs) of mRNAs, which allows for these factors to regulate various post-transcriptional processes, such as splicing, translation, and RNA stability. In fact, mRNAs are bound by an array of RBPs throughout their life cycle, positioning these factors to rapidly integrate cellular signals with changes in gene expression [4]. Additionally, RBPs often play essential roles in shaping the cellular response to stress conditions, particularly at the level of mRNA translation. For example, hypoxia reduces global translation rates via inactivation of eIF4E, but certain mRNAs involved in adaptation to low oxygen availability escape translational suppression under these conditions. This selective translation is dependent on the RBP RBM4 binding to 3′UTR regulatory elements to recruit translation machinery specifically to these transcripts [5]. Moreover, RBPs have been implicated in the cellular response to a range of stress conditions, including ER stress, DNA damage, oxidative stress, and viral infection [6], indicating that these factors are broadly important in cellular reprogramming during stress.
Despite these critical roles, our understanding of the RBPs mediating post-transcriptional regulation in β cells has significantly lagged behind that of transcriptional controls. One prominent example of RBP-mediated post-transcriptional regulation in β cells is the polypyrimidine tract binding protein (PTB). This RBP was found to stabilize mRNAs encoding components of secretory granules in response to high glucose levels, which enhances the insulin secretory capacity of the cell [7]. A growing body of work has highlighted several additional RBPs that control various processes in β cells via post-transcriptional regulation of gene expression [8].
In this study, we perform an unbiased search for post-transcriptional regulators in β cells, leading us to study the RBP hnRNPK, a member of the polyC-binding protein family of RBPs. hnRNPK is a highly multi-functional protein that can regulate many steps of gene expression, including translation [9]. Interestingly, this factor acts downstream of several signaling pathways, allowing for the integration of extracellular cues with changes in gene expression [10], [11], [12], [13]; however, the role of hnRNPK in β cells is unknown. By focusing on the post-transcriptional regulation of the mRNA encoding JUND, a transcription factor with links to β cell redox homeostasis and apoptosis [14], we elucidate a novel ERK/hnRNPK/DDX3X axis that is activated in islets during metabolic stress.
2. Material and methods
2.1. Animals
Animal studies were approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Wild type CD1 males were purchased from Jackson Laboratory. Male db/db mice (C57BLKS/J Leprdb/db) or db/+ mice (C57BLKS/J Leprdb/+) were purchased from Jackson Laboratory. Mice were housed in a 12hr light/dark cycle and had ad libitum access to food.
2.2. Lentivirus production
293T cells were transfected for 8 h in OptiMEM using Lipofectamine 2000 (Invitrogen), after which the media was changed to standard high glucose DMEM. psPAX2 and pMD2.G were used for packaging and envelope vectors. These plasmids were a gift from Didier Trono (Addgene plasmid #12260 and # 12259). Media containing virus was collected 2 and 3 days post-transfection. Ultracentrifugation of collected media (19,000rpm for 1.5 h at 4 °C) was used to concentrate virus. Lentivirus was titered by RT-PCR [15].
2.3. Cell line culture
Min6 mouse insulinoma cells passage 20–30 were cultured in high glucose (25 mM) DMEM as described [16], unless otherwise noted. For lentiviral infections, Min6 cells were transduced for 6 h with virus and polybrene (Sigma) at 8ug/mL. Cells were allowed to recover for 4–5 days before collection or stress treatments. HEK293T cells were cultured in DMEM containing 25 mM glucose.
2.4. GFP-RPL10A Min6 stable cell line
The GFP-RPL10A transgene was generated by cloning PCR amplified fragments for GFP-RPL10A or GFP into the pBABE-puro retroviral vector [17] digested with SalI. Retrovirus was produced in HEK293T cells and added to Min6 cells, followed by two rounds of puromycin selection (5 days, 2ug/mL).
2.5. Islet isolation and culture
Mouse islets were isolated from 6 to 12 week old CD1 male mice unless otherwise noted. Briefly, ductal inflation of the pancreas was performed followed by collagenase digestion (Roche 11213873001). Islets were enriched by density gradient centrifugation with Ficoll-Paque (GE 45-001-751). After handpicking 3–4 times, islets were collected for RNA/protein isolation or cultured overnight for recovery from isolation and stress treatments were started the next day.
Human islets were obtained through the NIH-supported Human Pancreas Analysis Program via the University of Pennsylvania Islet Core facility. The islets were harvested from non-diabetic deceased donors without any identifying information at NIH-approved centers with informed consent and IRB approval at the islet isolation centers. Human islet donor characteristics are provided in Supplementary Table 1.
The culture media used for mouse and human islets was RPMI 1640 (11 mM glucose) supplemented with 10% FBS, 2 mM glutamine, 1 mM sodium pyruvate, 10 mM HEPES, 1% antibiotic antimycotic (Thermo 15240096), and pH was adjusted to 7.3–7.4.
2.6. Palmitate preparation and glucolipotoxicity conditions
Palmitate (Sigma P9767) was dissolved in 50% ethanol at 65 °C and diluted in 10% BSA to a concentration of 7 mM. The mixture was incubated at 37 °C for 1 h to allow for conjugation before diluting in culturing media to a final concentration of 0.5 mM. Control media was made by performing the same procedure with 50% ethanol and no palmitate.
For islets, control media (described above) contained 11 mM glucose with no added palmitate. Media for glucolipotoxic conditions contained 25 mM glucose with 500uM palmitate.
For Min6, control media (DMEM) contained 5.6 mM glucose with no added palmitate while glucolipotoxic conditions had 25 mM glucose and 500uM palmitate.
2.7. RNA isolation and RT-qPCR
For Min6, cells were washed 2X with cold PBS before addition of TRIZOL (Invitrogen) and RNA was extracted according to manufacturer's instructions. RNA was reverse transcribed with random hexamers using High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). For islets, handpicked islets were washed 2X with cold PBS and RNA was extracted using RNeasy Mini Kit (Qiagen). RNA was reverse transcribed using oligo (dT) and Superscript III (Invitrogen). Quantitative PCR (BioRad CFX384) was used to measure transcript abundance and normalized to HPRT. See Supplementary Table 2 for a list of primer sequences used for these analyses.
2.8. Western blot
Proteins were separated by SDS-PAGE and immunoblotted with the following antibodies: rabbit anti-JUND (Santa Cruz, sc-74), rabbit anti-hnRNPK (Bethyl Laboratories, A300-674A), rabbit anti-DDX3X (Bethyl Laboratories, A300-474A), rabbit anti-phospho-ERK1/2 (Cell Signaling, 4377), rabbit anti-total-ERK1/2 (Cell Signaling, 4695), mouse anti-Tubulin (Sigma, T9026), and mouse anti-Ran (B.D. 610340).
2.9. Co-immunoprecipitation
Min6 cells or mouse islets were lysed in buffer containing 20 mM Tris pH 8.0, 137 mM NaCl, 1 mM MgCl2, 1 mM CaCl2, 1% NP-40, 10% glycerol, protease inhibitor cocktail (Millipore), and Benzonase (Sigma) at 12.5 U/mL. Lysates were rotated at 4 °C for 1 h. Protein concentration was determined using a Micro BCA Protein Assay Kit (Thermo). 1ug of primary antibody was added to lysate encompassing 500ug of protein and incubated overnight at 4 °C. Protein A Dynabeads were washed 3 times in lysis buffer then resuspended in lysate/antibody mixture and incubated for 3 h at 4 °C. The immunoprecipitations were washed 4 times with lysis buffer then eluted in NuPAGE LDS Sample Buffer (Invitrogen) by heating at 70 °C for 10 min. Eluted proteins and input samples were analyzed by western blot.
2.10. TRAP
TRAP was performed as described [18] with minor modifications. Briefly, after stress treatments of GFP-RPL10A Min6 cells, cycloheximide (Sigma) was added to the culture media at 100ug/mL for 10 min prior to washing 2X with cold PBS. Cells were lysed and protein concentration was measured by BCA (Thermo). Total RNA was isolated (1–5% input) with the RNeasy Mini Kit (Qiagen). For IP RNA, cell lysates encompassing 200ug of protein were added to Protein G Dynabeads bound to GFP antibodies (19C8 and 19F7, Memorial Sloan-Kettering Monoclonal Antibody Facility) and incubated overnight at 4 °C. The next day, the beads were washed 4X with high salt buffer, as described [18]. IP RNA was eluted from beads in RLT buffer and extracted using the RNeasy Mini Kit. Ribosome occupancy was determined by dividing transcript abundance for each gene in the IP RNA fraction by its level in the Total RNA fraction. For TRAP followed by RT-qPCR, RNA was reverse transcribed with random hexamers and Superscript III (Invitrogen) and transcript abundance was first normalized to HPRT for each fraction before determining ribosome occupancy.
2.11. Phos-tag analysis
Phos-tag was performed as described [19]. Briefly, lysates were run on SDS-PAGE gels containing 25uM Phos-tag acrylamide (Wako), 50uM MnCl2, and 8% acrylamide. The gels were run for 4 h at 90 V to achieve optimal separation of bands for hnRNPK. Prior to transfer, the gels were soaked in buffer containing 10 mM EDTA for 1 h.
2.12. RNA immunoprecipitation
RNA immunoprecipitation was performed using rabbit anti-hnRNPK (Bethyl Laboratories, A300-674A) or rabbit anti-DDX3X antibodies (Bethyl Laboratories, A300-474A). hnRNPK or DDX3X antibodies or IgG were bound to Protein A Dynabeads in buffer containing 20 mM HEPES pH 7.4, 5 mM MgCl2, 150 mM KCl, 2 mM DTT, 1% NP-40. Min6 cells or mouse islets were lysed in buffer containing 50 mM Tris pH 8.0, 150 mM NaCl, and 1% NP-40 with RNase, protease, and phosphatase inhibitors. Total RNA was extracted from lysate (10% input) using RNeasy Mini Kit (Qiagen). Lysate encompassing 75ug or 200ug of protein was added to beads bound to antibody for hnRNPK or DDX3X, respectively, and an IgG control, then incubated overnight at 4 °C. The next day, IPs were washed with buffer containing 20 mM HEPES pH 7.4, 5 mM MgCl2, 350 mM KCl, 2 mM DTT, 1% NP-40, and RNase inhibitors. RNA was eluted from beads in RLT buffer (Qiagen) and RNA was isolated using RNeasy Mini Kit (Qiagen). For RIP followed by RT-qPCR, enrichment was calculated by first normalizing transcript abundance in IP RNA to that in total RNA and then to the IgG control.
2.13. CRISPR design, cloning, and gene depletion
CRISPR gRNAs were designed using http://crispr.mit.edu/to minimize off-target binding (ROSA26: AAGATGGGCGGGAGTCTTCT, hnRNPK: GTTTAATACTTACGTCTGTA, DHX9: TTCAGTTGTGATTATCCGAG and GAGCGAGTTGCTTATGAGAG, DDX1: CAGATGAACCCATATGATAG and GGAACTAGAGGACTGCTGAA, DDX3X: AGATTGGATACTGTTTACGA and GCACCACCATAAACCACGCA). gRNAs were cloned into lentiCRISPR v2, as described [20].
CRISPR lentiviral particles were produced as described above and used to transduce Min6 cells. Cells were treated with puromycin (2ug/mL) for 3 days to select for transduced cells. Pooled cells were used for subsequent experiments and knockout was assessed by western blot.
2.14. MEK1-CA and hnRNPK cloning
Constitutively active MEK1 fragments were generated by PCR with primers to introduce S- > D mutations at Ser218 and Ser222. CA-MEK1 and the rat insulin promoter fragments were cloned into pLenti CMV blast [21] using Gibson Assembly (replaced CMV promoter with RIP).
hnRNPK was cloned into pLenti CMV blast along with the rat insulin promoter and an N-terminal HA tag using Gibson assembly.
2.15. Caspase-3 and-7 activation assay
Following stress treatment, 50–100 intact islets were transferred to a polysterene round bottom tube and fluorescent inhibitor of caspases (FLICA) reagent (Image-iT LIVE Red Caspase-3 and -7 Detect kit, Invitrogen) was added at 1:150 dilution followed by incubation at 37 °C for 1 h. Islets were washed 2X with wash buffer (provided by manufacturer), dispersed to single cell suspension, attached to slides using cytospin, and imaged using fluorescence microscopy (Keyence BZ-X700 microscope). Caspase positive cells were determined using BZ-X Advanced Analysis Software and colocalized with DAPI signal, and at least 200 cells were counted for each condition per experiment.
2.16. RIP-seq and analysis
RNA was isolated using the RNeasy Mini Kit (Qiagen) following RNA immunoprecipitation. Libraries were prepared using NEB Next Ultra RNA Library Prep Kit according to manufacturer's instructions with polyA enrichment followed by paired-end sequencing of 150bp using HiSeq (Illumina). Reads were mapped to mm10 using TopHat2 [22] and read counts per gene were determined using featureCounts [23]. edgeR was used to determine enrichment in the hnRNPK immunoprecipitated RNA compared to total RNA with significant genes called using a fold-change cutoff of greater than 1.5 and FDR less than 1 × 10−4. An IgG control was performed but not sequenced because no RNA was detected in the pull-down by Qubit RNA High Sensitivity quantitation. The overlap between RNA-seq data sets was determined using hypergeometric tests.
2.17. Statistics
Data are presented as mean ± standard error of the mean unless otherwise noted in figure legends. Statistical analyses were performed using GraphPad PRISM 7 software. Statistical tests used are noted in figure legends and include unpaired two-tailed Student's t-test, one-way analysis of variance (ANOVA), two-way ANOVA, and hypergeometric test.
2.18. Motif analysis
De novo motif discovery was performed using the MEME software suite [24] with the following parameters: strand-specific search, motif width 4–10. For motif frequency comparison, control sets of genes were generated by randomly selecting 100 genes 10 times from a list of genes with detectable expression in Min6 cells but not meeting the criteria for differential ribosome occupancy with PDX1 deficiency. The FIMO tool of the MEME suite [24] was used to determine motif frequencies. The Tomtom tool of the MEME suite [24] was used to compare discovered motifs to a database of motifs recognized by RBPs [25].
2.19. Data availability
RNA-seq data that support the findings of this study have been deposited in NCBI's GEO under accession codes GSE121296. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
3. Results
3.1. De novo motif analysis implicates the RNA binding protein hnRNPK in targeting the JUND mRNA in β cells
We previously used Translating Ribosome Affinity Purification followed by RNA-seq (TRAP-seq) to search for mRNAs with altered ribosome binding density in a PDX1-deficiency model of β cell dysfunction [14]. Depletion of PDX1 is a useful model to screen for factors with critical roles in β cells given that it is a human diabetes gene that shapes β cell susceptibility to stress [26], [27]. Indeed, this approach led us to uncover a subset of mRNAs regulated at the level of ribosome binding, including the mRNA encoding JUND, a transcription factor that promotes β cell apoptosis via regulation of pro-oxidant and pro-inflammatory genes [14]. In this study, we build on these findings by investigating the factors controlling JUND mRNA translation with the goal of uncovering novel pathways influencing β cell adaptation to stress conditions.
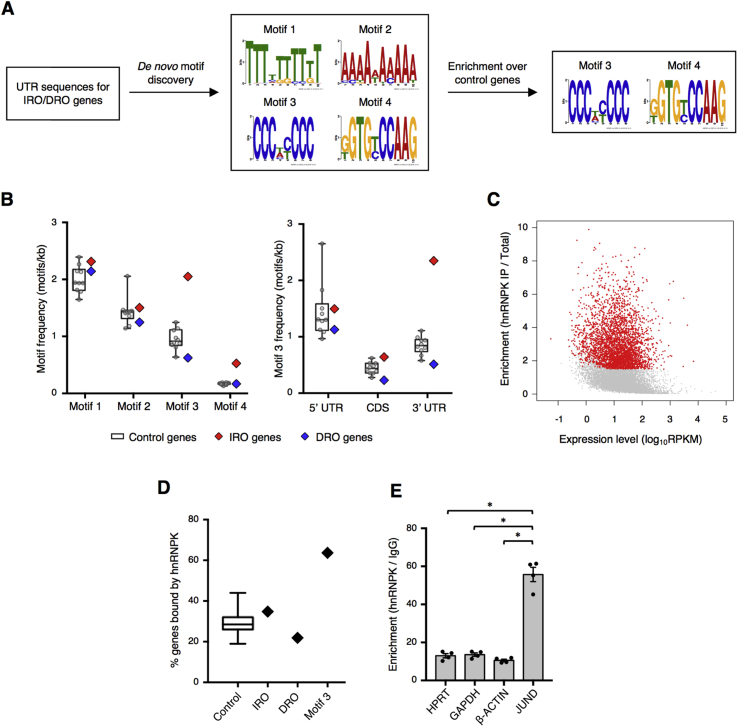
First, we used a bioinformatic approach to search for sequence motifs in our TRAP-seq data set, which included 53 genes with increased ribosome occupancy and 57 genes with decreased ribosome occupancy during PDX1-deficiency [14]. Specifically, we hypothesized that the genes with altered ribosome occupancy may share a common regulatory element in their untranslated regions (UTRs) that imparts translational regulation via RBP binding. Indeed, a de novo motif analysis using the MEME discovery tool [28] identified 4 motifs as occurring more frequently in UTRs of genes with increased ribosome occupancy than expected by chance (Figure 1A). However, this analysis will identify motifs that are generally enriched in UTRs in addition to those that are specific to the genes from our TRAP-seq data set. To distinguish between these possibilities, we randomly sampled 100 sets of 100 genes with no change in ribosome occupancy (control genes) for comparison of motif frequency (number of motifs per sequence length). Motifs 1 and 2 showed no enrichment over controls, indicating they are generally enriched in UTRs but not specifically enriched in the genes with altered ribosome occupancy (Figure 1A,B). In contrast, the motif frequency for the cytosine-rich motif 3 was significantly higher in the genes with increased ribosome occupancy compared to both control genes and genes with decreased ribosome occupancy, and it was specifically enriched in the 3′UTR of these genes (Figure 1A,B). Additionally, motif 3 was present in the 3′UTR of the transcript encoding JUND. Motif 4 was also enriched above controls, albeit at a lower frequency than motif 3, but was not present in the JUND UTRs (Figure 1B).
Figure 1.
Identification of hnRNPK as a mediator of post-transcriptional regulation in β cells. (A) Schematic demonstrating motif analysis on UTRs of genes with increased ribosome occupancy (IRO genes) or decreased ribosome occupancy (DRO genes) during PDX1-deficiency and motifs found by de novo motif discovery. (B) Assessment of motif frequencies in UTRs of genes with changes in ribosome occupancy after PDX1 depletion and sets of randomly sampled control genes. Box, 25–75th percentile; bar, median; whiskers, range. (C) Comparison of transcript expression level to its enrichment in the hnRNPK IP over total RNA as determined by RIP-seq for hnRNPK in Min6 cells treated with glucolipotoxicity. Genes with a statistically significant enrichment in hnRNPK binding are shown in red (n = 3). (D) Comparison of the percentage of genes enriched for hnRNPK binding by RIP-seq. Control group is 100 sets of 100 randomly sampled genes from all expressed genes. Box, 25–75th percentile; bar, median; whiskers, range. Increased ribosome occupancy (IRO) and decreased ribosome occupancy (DRO) are genes determined to have significant changes in ribosome occupancy by TRAP-seq during PDX1-deficiency. Motif 3 group is the set of IRO genes that contained at least one occurrence of motif 3 in the 3′ UTR. (E) Interaction of hnRNPK with the mRNA encoding JUND compared to housekeeping controls, as determined by RNA immunoprecipitation (n = 4). P values were calculated by unpaired two-tailed Student's t-test and * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
To investigate the significance of the cytosine-rich motif, we used MEME Suite tools to compare motif 3 to a database of motifs for RBPs [25] and found a significant alignment to the motifs recognized by the polyC-binding protein family of RBPs, including the member hnRNPK, a known regulator of mRNA translation [9] (Supplementary Fig. 1A). To determine whether genes from the TRAP screen were enriched for hnRNPK binding, RNA immunoprecipitation followed by RNA-seq (RIP-seq) for hnRNPK was performed in Min6 cells to assess hnRNPK-bound transcripts genome-wide (Figure 1C). Overlap of the RIP-seq and TRAP-seq data sets indicated that 64% of the genes with increased ribosome occupancy that contained at least one occurrence of motif 3 were enriched for hnRNPK binding. This constituted a statistically significant overlap between the two gene sets (Figure 1D, p = 1.0 × 10−4, hypergeometric test), indicating that the polyC-containing genes were enriched for hnRNPK binding.
To investigate the possibility that hnRNPK targets the JUND mRNA, we next examined the JUND 3′UTR sequence and found the presence of 23 and 40 bps of sequence matching the cytosine-rich motif in mouse and human, respectively (Supplementary Fig. 1B). Indeed, the JUND transcript was enriched for hnRNPK binding in our RIP-seq data, and we confirmed this interaction by RIP followed by RT-qPCR for hnRNPK in Min6 cells (Figure 1E). Analysis of hnRNPK eCLIP data in HepG2 and K562 cells [29] further confirmed this interaction and indicated that the binding occurs in the 3′UTR of JUND (Supplementary Fig. 1C). Thus, an unbiased search for UTR regulatory elements identified enrichment of a polyC motif and led to the finding that hnRNPK targets the JUND mRNA.
3.2. Post-transcriptional induction of JUND during metabolic stress is dependent on hnRNPK
Given the interaction between hnRNPK and the mRNA encoding JUND, we hypothesized that hnRNPK participates in the regulation of JUND expression. We have previously shown that JUND is post-transcriptionally upregulated in β cells during conditions of high glucose and free fatty acid, hereafter termed glucolipotoxicity, and that JUND promotes redox imbalance and apoptosis during these conditions [14]. Thus, we used the glucolipotoxicity model to further investigate the involvement of hnRNPK in JUND regulation. To confirm a post-transcriptional mechanism underlying the upregulation of JUND during metabolic stress, a time-course experiment was conducted, which showed that glucolipotoxic conditions led to a gradual increase in JUND protein levels over time with no increase in JUND mRNA abundance at any time-point (Supplementary Fig. 2).
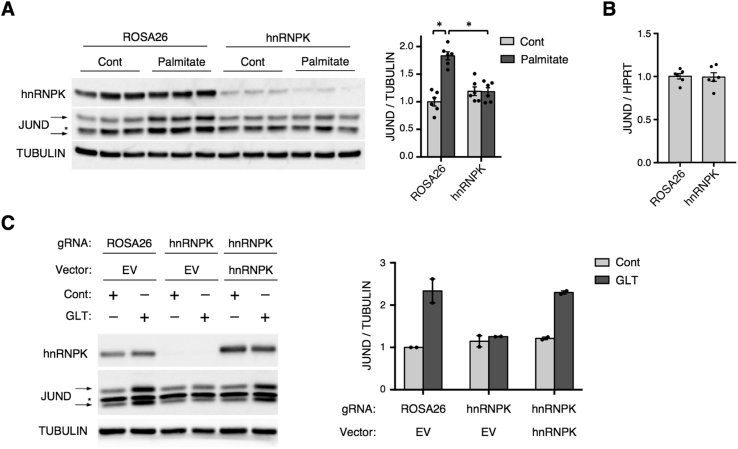
We next investigated whether hnRNPK is required for the upregulation of JUND during metabolic stress using CRISPR-mediated targeting to robustly deplete hnRNPK in Min6 cells compared to control cells receiving a gRNA targeting the ROSA26 locus (Figure 2A). As expected, treatment of the ROSA26 group with high levels of palmitate led to increased JUND protein levels (Figure 2A). Strikingly, depletion of hnRNPK completely blocked this induction of JUND despite having no impact on JUND mRNA abundance (Figure 2A,B). To confirm that this effect was specific for loss of hnRNPK, we used a CRISPR-resistant construct to rescue expression of hnRNPK in cells that also expressed a gRNA targeting the endogenous hnRNPK gene. Indeed, overexpression of hnRNPK restored the upregulation of JUND during glucolipotoxicity to the same level as the ROSA26 group (Figure 2C). Together, these data indicate that hnRNPK regulates the post-transcriptional induction of JUND during metabolic stress.
Figure 2.
hnRNPK is required for JUND induction during metabolic stress. (A) Western blot and quantification showing CRISPR-mediated depletion of hnRNPK in Min6 cells blocks the induction of JUND after 2 days of palmitate treatment (n = 6). (B) No change in JUND transcript levels with depletion of hnRNPK in Min6 cells during palmitate treatment, as determined by RT-qPCR (n = 6). (C) Western blot and quantification showing depletion of hnRNPK in Min6 cells using CRISPR-Cas9 and rescue of hnRNPK expression with a CRISPR-resistant construct. Control groups include a gRNA targeting the ROSA26 locus and the empty vector (EV) expression plasmid. Cells were treated for 30hrs in control (Cont) or glucolipotoxic (GLT) conditions (n = 2). P values were calculated by two-way ANOVA in (A) or unpaired two-tailed Student's t-test in (B). For Western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
3.3. hnRNPK is phosphorylated in β cells during metabolic stress
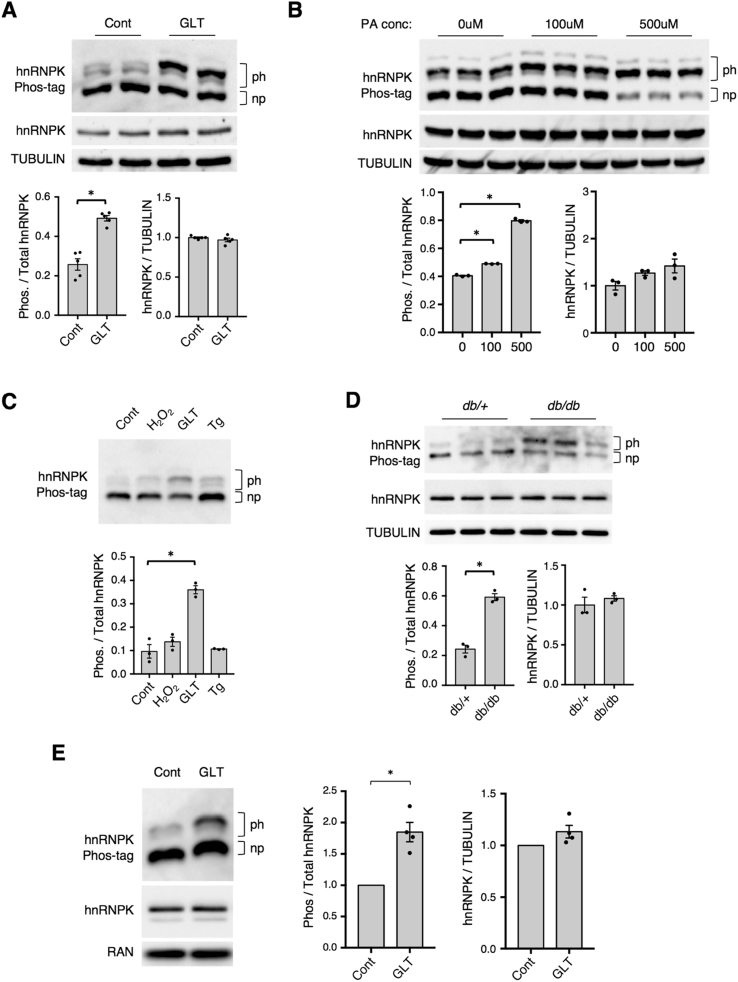
While loss of hnRNPK prevented the upregulation of JUND during metabolic stress, it did not impact JUND levels under control conditions (Figure 2A,C), suggesting that the function of hnRNPK may be altered in a stress-dependent manner. A common mechanism for regulation of hnRNPK function is post-translational modification via phosphorylation [9]. To study changes in hnRNPK phosphorylation, we used Phos-tag SDS-PAGE, which unveils phosphorylated forms of hnRNPK via electrophoresis [30]. This approach clearly demonstrated a shift towards increased phosphorylation of hnRNPK in both mouse islets and Min6 cells exposed to metabolic stress (Figure 3A,B). This increase in phosphorylation was specific for metabolic stress, as it did not occur during stress induced by hydrogen peroxide or thapsigargin (Figure 3C). Similarly, there was an increase in hnRNPK phosphorylation in isolated islets from diabetic db/db mice and in human islets treated with glucolipotoxicity, indicating that this pathway is also activated during metabolic stress in vivo and is conserved in human islets (Figure 3D,E). Further there was no change in total hnRNPK levels during glucolipotoxicity, suggesting that hnRNPK is primarily regulated at the level of post-translational modification under these conditions (Figure 3). Importantly, we previously observed an increase in JUND levels in all of these models of metabolic stress [14], indicating a robust association between JUND induction and hnRNPK phosphorylation.
Figure 3.
hnRNPK is phosphorylated in mouse and human islets and Min6 cells during metabolic stress. Phos-tag SDS-PAGE analysis shows changes in phosphorylation of hnRNPK in (A) mouse islets treated with glucolipotoxic (GLT) conditions for 2 days (n = 5), (B) Min6 cells treated with 100uM or 500uM of palmitate for 2 days (n = 3), (C) mouse islets treated with a panel of stressors including hydrogen peroxide (H2O2, 200uM) for 1 h, GLT for 2 days, or thapsigargin (Tg, 1uM) for 3 h (n = 3), (D) islets from 12-wk old db/db or db/+ male mice (n = 3), and (E) human islets treated with glucolipotoxic (GLT) conditions for 2 days (n = 4). np denotes band for non-phosphorylated hnRNPK and ph denotes bands for phosphorylated hnRNPK. Quantification of Phos-tag blots represents the signal intensity of the phosphorylated bands divided by the sum of the signal intensities for all bands (non-phosph and phosph). Results from standard SDS-PAGE analyses for hnRNPK and a loading control are depicted below Phos-tag gels to quantify total hnRNPK levels. P values were calculated by unpaired two-tailed Student's t-test and * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
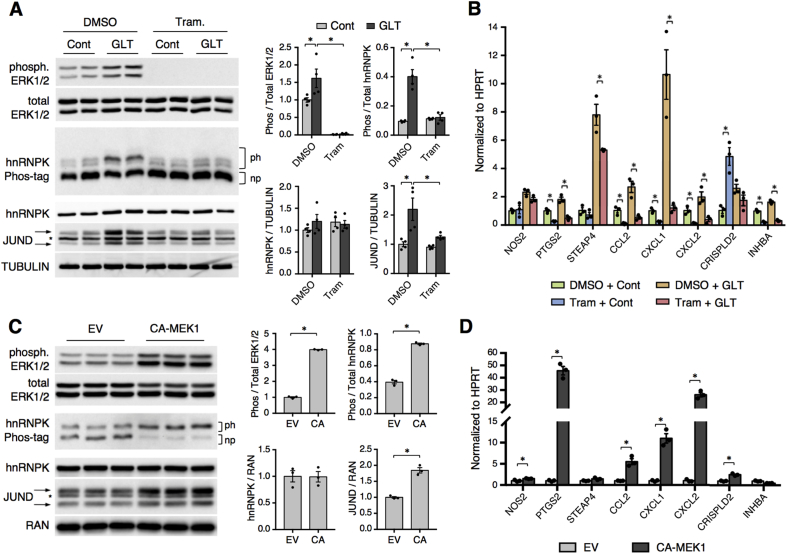
3.4. The MEK/ERK signaling pathway is necessary and sufficient to activate the hnRNPK/JUND pathway
One of the kinase cascades that targets hnRNPK and modulates its function is the MEK/ERK signaling pathway [10], which was activated in mouse islets treated with high levels of glucose and palmitate (Figure 4A). Treatment of mouse islets with the potent and specific MEK1/2 inhibitor trametinib during metabolic stress completely abolished p-ERK1/2 levels, confirming successful pathway inhibition. Interestingly, MEK inhibition also blocked the increase in phosphorylation of hnRNPK and the induction of JUND in islets treated with glucolipotoxicity (Figure 4A). Further, trametinib treatment of mouse islets reduced expression levels for most of the genes previously identified as JUND targets in β cells [14], including Ptgs2 and Steap4 (Figure 4B). Of note, MEK inhibition reduced the expression of several targets under both control and glucolipotoxic conditions, indicating that MEK/ERK signaling likely impacts the expression of these genes by both JUND-dependent and –independent mechanisms. Together, these data indicate that MEK/ERK signaling is necessary for the activation of the hnRNPK/JUND pathway during metabolic stress. To test the sufficiency of ERK to activate the hnRNPK/JUND pathway, we overexpressed a constitutively active form of MEK1 (S218/222D) in Min6 cells. As expected, this led to a robust increase in phosphorylation of ERK1/2 (Figure 4C). Consistent with the effect of MEK inhibition, expression of constitutively active MEK1 was sufficient to increase hnRNPK phosphorylation, JUND levels, and expression of many JUND targets, most notably Ptgs2 (Figure 4D).
Figure 4.
The MEK/ERK signaling pathway is necessary and sufficient for activation of the hnRNPK/JUND pathway. (A) Western blot and Phos-tag SDS-PAGE analyses show that trametinib treatment (1uM) blocks ERK phosphorylation and activation of the hnRNPK/JUND pathway in mouse islets cultured for 2 days in control (Cont) or glucolipotoxic (GLT) conditions (n = 4). (B) RT-qPCR analysis for JUND targets in mouse islets treated with trametinib (1uM) and cultured for 2 days in control or GLT conditions (n = 3). (C) Lentiviral overexpression of constitutively active (CA) MEK1 (S218/222D) in Min6 cells activates the ERK/hnRNPK/JUND pathway as determined by western blot and Phos-tag SDS-PAGE (n = 3). (D) RT-qPCR analysis for JUND targets in Min6 cells with overexpression of CA-MEK1 (n = 3). EV denotes lentiviral transduction with empty vector. P values were calculated by two-way ANOVA (A,B) or unpaired two-tailed Student's t-test (C,D). For western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
Given our previous findings that depletion of JUND reduced apoptosis in β cells during metabolic stress [14], we reasoned that inhibiting the MEK/ERK signaling pathway with trametinib may also improve cell survival during glucolipotoxicity. To test this possibility, mouse islets were cultured in glucolipotoxic conditions with or without trametinib, and apoptosis was measured using a fluorescence readout of caspase activation. Strikingly, MEK inhibition provided a significant decrease in islet cell apoptosis during metabolic stress (Supplementary Fig. 3), consistent with the effect of JUND depletion. Notably, metabolic stress predominantly causes apoptosis of β cells rather than α cells [31], indicating that MEK inhibition likely plays a protective role in β cells.
3.5. The RNA helicase DDX3X interacts with hnRNPK and is required for the efficient translation of JUND mRNA
To explore the mechanism by which hnRNPK impacts JUND translation during metabolic stress, we considered several possibilities based on modifiable properties of hnRNPK, including its subcellular localization, binding affinity for RNA, and nuclear retention of mRNA [10], [11], [32]. We were unable to detect changes in these features during glucolipotoxicity that could explain the induction of JUND (Supplementary Fig. 4); therefore, we considered the possibility that hnRNPK impacts JUND translation via interaction with a cofactor. Interestingly, the JUND mRNA has been reported to contain a highly structured 5′UTR [33], and efficient translation initiation is dependent on RNA helicases, such as DHX9, to resolve these secondary structures [34]. Thus, we hypothesized that hnRNPK may interact with an RNA helicase that enhances the translation of JUND mRNA. Upon searching mass spectrometry data sets for hnRNPK interacting factors [35], [36], we noticed the presence of the DEAD-box helicases DHX9, DDX1, and DDX3X in the hnRNPK interactome. Given the connection of DEAD-box helicases to the regulation of translation [34], [37], we selected these three genes as our top candidates and assessed their impact on JUND expression by using CRISPR-Cas9 to efficiently deplete these genes in Min6 cells (Figure 5A,B). Interestingly, loss of DDX3X, but not DHX9 nor DDX1, led to reduced JUND protein levels in Min6 cells during glucolipotoxicity (Figure 5C). Notably, loss of DDX3X led to a significant increase in JUND mRNA levels (Figure 5D). The decrease in the amount of JUND protein despite an increase in mRNA abundance likely signifies a defect in translation initiation caused by loss of DDX3X, consistent with its ability to enhance translation of select mRNAs [38].
Figure 5.
The RNA helicase DDX3X is required for efficient translation of JUND mRNA. (A) RT-qPCR for DHX9, DDX1, and DDX3X and (B) western blot for DDX3X show a significant depletion of these RNA helicases using CRISPR-Cas9 in Min6 cells (n = 3). (C) Representative western blot depicting JUND levels in Min6 cells transduced with lentivirus to deplete the indicated RNA helicase using CRISPR-Cas9 and cultured in glucolipotoxic conditions for 30hrs (n = 3). (D) Assessment of JUND mRNA abundance in Min6 cells by RT-qPCR after CRISPR-mediated depletion of DDX3X and culturing in glucolipotoxic conditions for 30hrs (n = 3). (E) Decreased ribosome occupancy of JUND in GFP-RPL10a Min6 cells after depletion of DDX3X by CRISPR-Cas9 and 30 h of glucolipotoxic conditions, as determined by TRAP followed by RT-qPCR (n = 3). * = p < 0.05 and ns = not significant. P values were calculated by two-way ANOVA except in (B) in which an unpaired two-tailed Student's t-tests was used. In (D), * denotes significance compared to ROSA26 group of the same treatment unless otherwise noted. For western blot images of JUND, arrows denote two bands for JUND and * denotes a non-specific band. Otherwise, * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
Indeed, depletion of DDX3X in GFP-RPL10A Min6 cells led to a significant reduction in the density of ribosomes binding to the JUND mRNA, as determined by TRAP (Figure 5E). These data indicate that DDX3X is required for the efficient translation of JUND mRNA in Min6 cells.
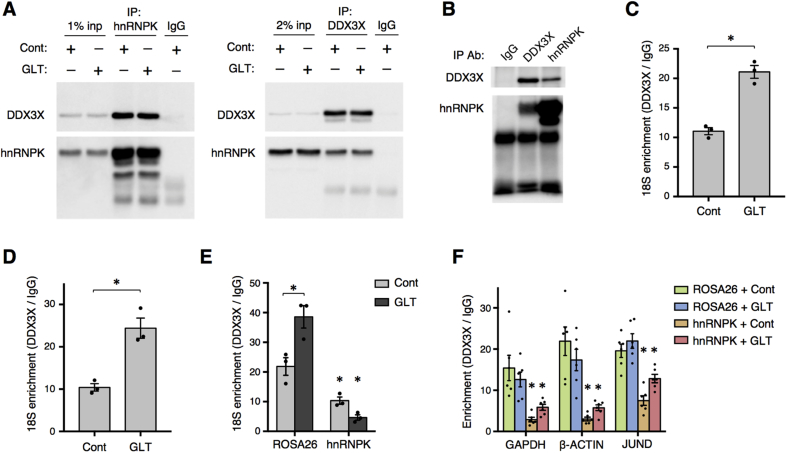
To assess the interaction between hnRNPK and DDX3X, we performed co-immunoprecipitation experiments in the presence of a nuclease to degrade both RNA and DNA. This showed a clear nucleic acid-independent interaction between these two factors that was maintained during glucolipotoxicity in Min6 cells (Figure 6A). Furthermore, co-immunoprecipitation experiments in mouse islets demonstrated that these factors also interact in primary tissue (Figure 6B). DDX3X and its yeast homolog Ded1 can impact translation through interaction with components of the 43S translation pre-initiation complex (PIC), especially the 18S ribosomal RNA [39], [40]. Indeed, RIP for DDX3X showed a robust binding to 18S in Min6 cells, and this interaction was increased during metabolic stress (Figure 6C). Importantly, the DDX3X-18S interaction occurs in primary mouse islets and is increased by glucolipotoxicity (Figure 6D). These findings indicate that DDX3X interacts with translation machinery in a stress-dependent manner, which may contribute to its effect on JUND translation. To test whether the DDX3X-18S interaction was influenced by hnRNPK, we performed RIP for DDX3X in Min6 cells with CRISPR-mediated depletion of hnRNPK. Intriguingly, loss of hnRNPK significantly reduced the association between DDX3X and 18S (Figure 6E). DDX3X has been reported to bind to all expressed mRNAs, likely via its interaction with the 43S PIC [39], [40]. Consistent with this, DDX3X bound to all analyzed mRNAs by RIP, and these interactions were also significantly reduced with loss of hnRNPK (Figure 6F). Together, these data support a model whereby hnRNPK impacts translation in part by promoting the interaction between DDX3X and the 43S PIC. Thus, we have identified DDX3X as a novel regulator of JUND translation during metabolic stress that interacts with the 18S ribosomal RNA in an hnRNPK-dependent manner.
Figure 6.
hnRNPK interacts with DDX3X and promotes DDX3X-18S interaction. (A) Western blots from pull-down of hnRNPK (left) or DDX3X (right) in Min6 cells treated with control (Cont) or glucolipotoxic (GLT) conditions for 30 h. (B) Representative Western blot from pull-down of hnRNPK and DDX3X in mouse islets indicating interaction between these factors in primary tissue. (C) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in Min6 cells treated with Cont or GLT conditions for 30hrs (n = 3). (D) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in mouse islets treated with Cont or GLT conditions for 2 days (n = 3). (E) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in Min6 cells with CRISPR-mediated depletion of hnRNPK and cultured in Cont or GLT conditions for 30hrs (n = 3). (F) Decreased interaction between DDX3X and indicated mRNAs after CRISPR-mediated depletion of hnRNPK in Min6 cells treated with Cont or GLT conditions for 30hrs as determined by RNA immunoprecipitation for DDX3X (n = 3). P values were calculated by two-way ANOVA except in (C) and (D) in which unpaired two-tailed Student's t-tests were used. In (E,F), * denotes significance compared to ROSA26 group of the same treatment unless otherwise noted. * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.
4. Discussion
Although the transcriptional regulators that shape the proper β cell gene expression program have received much attention, the factors important for post-transcriptional regulation in β cells, such as RBPs, are less defined but may be critical for adaptation to environmental stressors. In this study, an unbiased search for putative regulatory motifs led to the identification of hnRNPK as a post-transcriptional regulator of JUND expression. We show for the first time that hnRNPK is phosphorylated in β cells during conditions associated with T2D, and MEK/ERK signaling is both necessary and sufficient for hnRNPK phosphorylation and JUND induction. We identify the RNA helicase DDX3X as an hnRNPK-interacting factor that promotes the efficient translation of JUND mRNA. Given the pro-apoptotic role for JUND in β cells, these mechanistic findings uncover new possibilities for therapeutic intervention in T2D. Indeed, we find that MEK inhibition significantly reduces apoptosis in islets during metabolic stress.
The mechanisms by which hnRNPK influences multiple steps of gene expression are poorly understood but likely involve interactions with a range of cofactors [9]. In this study, we show that DDX3X and hnRNPK interact and converge on the post-transcriptional regulation of JUND expression. We further link these factors by showing that loss of hnRNPK reduces the association of DDX3X with 18S ribosomal RNA. It is currently unclear how hnRNPK impacts the DDX3X-18S interaction. One possibility is that hnRNPK facilitates this interaction via assembly of a large complex that includes DDX3X and the 43S PIC, and the composition of this complex may be altered during metabolic stress in an hnRNPK- and ERK-dependent manner. Additionally, this complex may contain cofactors that modulate DDX3X activity, as has been reported for EZRIN and GLE1 [41], [42]. Thus, it will be worthwhile to comprehensively define the hnRNPK and DDX3X interactome in β cells under control and glucolipotoxic conditions to identify other components of this hnRNPK-DDX3X complex and to determine whether they are recruited in a stress-dependent manner.
hnRNPK can be phosphorylated downstream of multiple signaling pathways, including Protein kinase C, JNK, and ERK [10], [43]. In β cells, we have found that activation of the MEK/ERK signaling pathway is both necessary and sufficient to increase hnRNPK phosphorylation and induce JUND levels. Furthermore, in the context of metabolic stress, inhibition of this ERK/hnRNPK/DDX3X axis with trametinib reduced apoptosis in mouse islets. Given that activation of MEK/ERK signaling will likely influence numerous cellular processes in β cells, it is currently unclear to what extent this effect on apoptosis can be attributed to modulation of hnRNPK/JUND. The pro-survival effect of trametinib during glucolipotoxicity is consistent with previous findings that MEK inhibition reduced apoptosis in human islets treated with high glucose levels or IL-1β [44]. In contrast, activation of the MEK/ERK signaling pathway has been shown to promote β cell survival in the context of incretin signaling [45], [46]. Thus, the impact of MEK inhibition on β cell viability is context-dependent. Interestingly, treatment of ob/ob mice with trametinib improves glucose homeostasis in these animals, however, this effect appears to be largely attributable to an improvement in insulin sensitivity [47]. Thus, the use of MEK inhibition in metabolic syndrome warrants further investigation for improving both insulin sensitivity and β cell viability.
hnRNPK is a highly multi-functional protein and will likely have diverse roles in β cells, including the regulation of transcription, splicing, and mRNA stability [9]. This versatility is intriguing in that hnRNPK can integrate signaling pathways with multiple aspects of RNA processing, but it also complicates the design and interpretation of loss-of-function studies. Nevertheless, given its phosphorylation during metabolic stress in vivo, a systematic analysis of hnRNPK function holds promise to elucidate novel aspects of β cell biology. Here, by focusing on hnRNPK interacting factors, we identified the RNA helicase DDX3X as a novel hnRNPK partner in the regulation of JUND translation. While the function of DDX3X in β cells is unknown, its links to cell cycle progression, apoptosis, and innate immunity in other cell types [48] warrant a broader examination of DDX3X function in β cells
Finally, it is worth noting that although our motif analysis suggests that the polyC stretches in the JUND 3′UTR impart its post-transcriptional regulation, we have been unable to directly test this possibility. Specifically, attempts to model JUND regulation using several reporter systems that contain the JUND UTRs did not recapitulate the regulation of endogenous JUND during glucolipotoxicity. It is unclear why these artificial reporters are insufficient to model JUND regulation, but may involve changes to RNA secondary structures or unidentified additional features of the JUND locus. Therefore, while we have demonstrated that hnRNPK directly targets the JUND mRNA, we cannot rule out the possibility that hnRNPK impacts JUND expression independent of polyC motif binding.
In conclusion, by focusing on the post-transcriptional regulation of JUND, we have uncovered a novel ERK/hnRNPK/DDX3X pathway in β cells that influences mRNA translation during conditions associated with T2D. Our identification of this pathway opens up new areas of investigation that promise to advance our understanding of β cell failure in T2D.
Author contributions
A.L.G. conceived of, designed, and performed experiments, interpreted results, and drafted and reviewed the manuscript. M.W.H. designed and performed experiments. N.M.D. and A.U.O. performed experiments. D.A.S. conceived of and designed the studies, interpreted results, and edited and reviewed the manuscript.
Acknowledgments
We thank the University of Pennsylvania Diabetes Research Center (DRC) for the use of the Functional Genomics Core and the Mouse Phenotyping, Physiology and Metabolism Core (P30-DK19525). Specifically, we acknowledge Dr. A. Rozo and A. Gonzalez for assistance with db/db mouse dissections. We also thank Dr. S. Liebhaber and Dr. K. Lynch for helpful discussions. This work was supported by National Institutes of Health Grants F30-DK105758 (to A.L.G.) and P01-DK49210 (to D.A.S.). We also would like to thank the Medical Scientist Training Program (MSTP) at the University of Pennsylvania for their support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2019.05.009.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Prentki M., Nolan C.J. Islet β cell failure in type 2 diabetes. Journal of Clinical Investigation. 2006;116(7):1802–1812. doi: 10.1172/JCI29103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Talchai C., Xuan S., Lin H.V., Sussel L., Accili D. Pancreatic β cell dedifferentiation as a mechanism of diabetic β cell failure. Cell. 2012;150(6):1223–1234. doi: 10.1016/j.cell.2012.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim-Muller J.Y., Fan J., Kim Y.J.R., Lee S.-A., Ishida E., Blaner W.S. Aldehyde dehydrogenase 1a3 defines a subset of failing pancreatic β cells in diabetic mice. Nature Communications. 2016;7:1–11. doi: 10.1038/ncomms12631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell S.F., Parker R. Principles and properties of eukaryotic mRNPs. Molecular Cell. 2014;54(4):547–558. doi: 10.1016/j.molcel.2014.04.033. [DOI] [PubMed] [Google Scholar]
- 5.Uniacke J., Holterman C.E., Lachance G., Franovic A., Jacob M.D., Fabian M.R. An oxygen-regulated switch in the protein synthesis machinery. Nature. 2012;486(7401):126–129. doi: 10.1038/nature11055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harvey R., Dezi V., Pizzinga M., Willis A.E. Post-transcriptional control of gene expression following stress: the role of RNA-binding proteins. Biochemical Society Transactions. 2017;45(4):1007–1014. doi: 10.1042/BST20160364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knoch K.-P., Bergert H., Borgonovo B., Saeger H.-D., Altkrüger A., Verkade P. Polypyrimidine tract-binding protein promotes insulin secretory granule biogenesis. Nature Cell Biology. 2004;6(3):207–214. doi: 10.1038/ncb1099. [DOI] [PubMed] [Google Scholar]
- 8.Magro M.G., Solimena M. Regulation of β-cell function by RNA-binding proteins. Molecular Metabolism. 2013;2(4):348–355. doi: 10.1016/j.molmet.2013.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bomsztyk K., Denisenko O., Ostrowski J. hnRNP K: one protein multiple processes. BioEssays. 2004;26(6):629–638. doi: 10.1002/bies.20048. [DOI] [PubMed] [Google Scholar]
- 10.Habelhah H., Shah K., Huang L., Ostareck-Lederer A., Burlingame A.L., Shokat K.M. ERK phosphorylation drives cytoplasmic accumulation of hnRNP-K and inhibition of mRNA translation. Nature Cell Biology. 2001;3(3):325–330. doi: 10.1038/35060131. [DOI] [PubMed] [Google Scholar]
- 11.Ostrowski J., Kawata Y., Schullery D.S., Denisenko O.N., Higaki Y., Abrass C.K. Insulin alters heterogeneous nuclear ribonucleoprotein K protein binding to DNA and RNA. Proceedings of the National Academy of Sciences. 2001;98(16):9044–9049. doi: 10.1073/pnas.161284098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostareck-Lederer A., Ostareck D.H., Cans C., Neubauer G., Bomsztyk K., Superti-Furga G. c-Src-mediated phosphorylation of hnRNP K drives translational activation of specifically silenced mRNAs. Molecular and Cellular Biology. 2002;22(13):4535–4543. doi: 10.1128/MCB.22.13.4535-4543.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchins E.J., Szaro B.G. c-Jun N-terminal kinase phosphorylation of heterogeneous nuclear ribonucleoprotein K regulates vertebrate axon outgrowth via a posttranscriptional mechanism. Journal of Neuroscience. 2013;33(37):14666–14680. doi: 10.1523/JNEUROSCI.4821-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Good A.L., Cannon C.E., Haemmerle M.W., Yang J., Stanescu D.E., Doliba N.M. JUND regulates pancreatic β cell survival during metabolic stress. Molecular Metabolism. 2019;25:95–106. doi: 10.1016/j.molmet.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sastry L., Johnson T., Hobson M.J., Smucker B., Cornetta K. Titering lentiviral vectors: comparison of DNA, RNA and marker expression methods. Gene Therapy. 2002;9(17):1155–1162. doi: 10.1038/sj.gt.3301731. [DOI] [PubMed] [Google Scholar]
- 16.Claiborn K.C., Sachdeva M.M., Cannon C.E., Groff D.N., Singer J.D., Stoffers D.A. Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. Journal of Clinical Investigation. 2010;120(10):3713–3721. doi: 10.1172/JCI40440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morgenstern J.P., Land H. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Research. 1990;18(12):3587–3596. doi: 10.1093/nar/18.12.3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle J.P., Dougherty J.D., Heiman M., Schmidt E.F., Stevens T.R., Ma G. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135(4):749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kinoshita E., Kinoshita-Kikuta E., Takiyama K., Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Molecular & Cellular Proteomics – MCP. 2006;5(4):749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Sanjana N.E., Shalem O., Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nature Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campeau E., Ruhl V.E., Rodier F., Smith C.L., Rahmberg B.L., Fuss J.O. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4(8):e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Bailey T.L., Boden M., Buske F.A., Frith M., Grant C.E., Clementi L. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Research. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. Web Server issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ray D., Kazan H., Cook K.B., Weirauch M.T., Najafabadi H.S., Li X. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2014;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stoffers D.A., Ferrer J., Clarke W.L., Habener J.F. Early-onset type-ll diabetes mellitus (MODY4) linked to IPF1. Nature Genetics. 1997;17(2):138–139. doi: 10.1038/ng1097-138. [DOI] [PubMed] [Google Scholar]
- 27.Sachdeva M.M., Sachdeva M.M., Claiborn K.C., Claiborn K.C., Khoo C., Khoo C. Pdx1 (MODY4) regulates pancreatic beta cell susceptibility to ER stress. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(45):19090–19095. doi: 10.1073/pnas.0904849106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings International Conference on Intelligent Systems for Molecular Biology. 1994;2:28–36. [PubMed] [Google Scholar]
- 29.The ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;488(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura Y., Nagata K., Suzuki N., Yokoyama R., Yamanaka Y., Kitamura H. Characterization of multiple alternative forms of heterogeneous nuclear ribonucleoprotein K by phosphate-affinity electrophoresis. Proteomics. 2010;10(21):3884–3895. doi: 10.1002/pmic.201000349. [DOI] [PubMed] [Google Scholar]
- 31.Marroqui L., Masini M., Merino B., Grieco F.A., Millard I., Dubois C. Pancreatic α cells are resistant to metabolic stress-induced apoptosis in type 2 diabetes. EBioMedicine. 2015;2(5):378–385. doi: 10.1016/j.ebiom.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lubelsky Y., Ulitsky I. Sequences enriched in Alu repeats drive nuclear localization of long RNAs in human cells. Nature. 2018;555(7694):107–111. doi: 10.1038/nature25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Short J.D., Pfarr C.M. Translational regulation of the JunD messenger RNA. Journal of Biological Chemistry. 2002;277(36):32697–32705. doi: 10.1074/jbc.M204553200. [DOI] [PubMed] [Google Scholar]
- 34.Hartman T.R., Qian S., Bolinger C., Fernandez S., Schoenberg D.R., Boris-Lawrie K. RNA helicase A is necessary for translation of selected messenger RNAs. Nature Structural & Molecular Biology. 2006;13(6):509–516. doi: 10.1038/nsmb1092. [DOI] [PubMed] [Google Scholar]
- 35.Mikula M., Rubel T., Karczmarski J., Statkiewicz M., Bomsztyk K., Ostrowski J. Beads-free protein immunoprecipitation for a mass spectrometry-based interactome and posttranslational modifications analysis. Proteome Science. 2015:1–7. doi: 10.1186/s12953-015-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H.-C., Lin W.-C., Tsay Y.-G., Lee S.-C., Chang C.-J. An RNA helicase, DDX1, interacting with poly(A) RNA and heterogeneous nuclear ribonucleoprotein K. Journal of Biological Chemistry. 2002;277(43):40403–40409. doi: 10.1074/jbc.M206981200. [DOI] [PubMed] [Google Scholar]
- 37.Linder P., Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nature Publishing Group. 2011;12(8):505–516. doi: 10.1038/nrm3154. [DOI] [PubMed] [Google Scholar]
- 38.Soto-Rifo R., Rubilar P.S., Limousin T., de Breyne S., cimo D.D.E., Ohlmann T.E.O. DEAD-box protein DDX3 associates with eIF4F to promote translation of selected mRNAs. The EMBO Journal. 2012;31(18):3745–3756. doi: 10.1038/emboj.2012.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oh S., Flynn R.A., Floor S.N., Purzner J., Martin L., Do B.T. Medulloblastoma-associated DDX3 variant selectively alters the translational response to stress. Oncotarget. 2016;7(19):28169–28182. doi: 10.18632/oncotarget.8612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guenther U.-P., Weinberg D.E., Zubradt M.M., Tedeschi F.A., Stawicki B.N., Zagore L.L. The helicase Ded1p controls use of near-cognate translation initiation codons in 5′ UTRs. Nature. 2018:1–20. doi: 10.1038/s41586-018-0258-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Çelik H., Sajwan K.P., Selvanathan S.P., Marsh B.J., Pai A.V., Saygideger Kont Y. Ezrin binds to DEAD-box RNA helicase DDX3 and regulates its function and protein level. Molecular and Cellular Biology. 2015;107:15–18. doi: 10.1128/MCB.00332-15. MCB.00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bolger T.A., Wente S.R. Gle1 is a multifunctional DEAD-box protein regulator that modulates Ded1 in translation initiation. Journal of Biological Chemistry. 2011;286(46):39750–39759. doi: 10.1074/jbc.M111.299321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schullery D.S., Ostrowski J., Denisenko O.N., Stempka L., Shnyreva M., Suzuki H. Regulated interaction of protein kinase Cdelta with the heterogeneous nuclear ribonucleoprotein K protein. Journal of Biological Chemistry. 1999;274(21):15101–15109. doi: 10.1074/jbc.274.21.15101. [DOI] [PubMed] [Google Scholar]
- 44.Maedler K., Størling J., Sturis J., Zuellig R.A., Spinas G.A., Arkhammar P.O.G. Glucose- and interleukin-1beta-induced beta-cell apoptosis requires Ca2+ influx and extracellular signal-regulated kinase (ERK) 1/2 activation and is prevented by a sulfonylurea receptor 1/inwardly rectifying K+ channel 6.2 (SUR/Kir6.2) selective potassium channel opener in human islets. Diabetes. 2004;53(7):1706–1713. doi: 10.2337/diabetes.53.7.1706. [DOI] [PubMed] [Google Scholar]
- 45.Quoyer J., Longuet C., Broca C., Linck N., Costes S., Varin E. GLP-1 mediates antiapoptotic effect by phosphorylating Bad through a beta-arrestin 1-mediated ERK1/2 activation in pancreatic beta-cells. Journal of Biological Chemistry. 2010;285(3):1989–2002. doi: 10.1074/jbc.M109.067207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Campbell J.E., Ussher J.R., Mulvihill E.E., Kolic J., Baggio L.L., Cao X. TCF1 links GIPR signaling to the control of beta cell function and survival. Nature Medicine. 2016;22(1):84–90. doi: 10.1038/nm.3997. [DOI] [PubMed] [Google Scholar]
- 47.Banks A.S., McAllister F.E., Camporez J.P.G., Zushin P.-J.H., Jurczak M.J., Laznik-Bogoslavski D. An ERK/Cdk5 axis controls the diabetogenic actions of PPARγ. Nature. 2015;517(7534):391–395. doi: 10.1038/nature13887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ariumi Y. Multiple functions of DDX3 RNA helicase in gene regulation, tumorigenesis, and viral infection. Frontiers in Genetics. 2014;5(1664):423. doi: 10.3389/fgene.2014.00423. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq data that support the findings of this study have been deposited in NCBI's GEO under accession codes GSE121296. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.