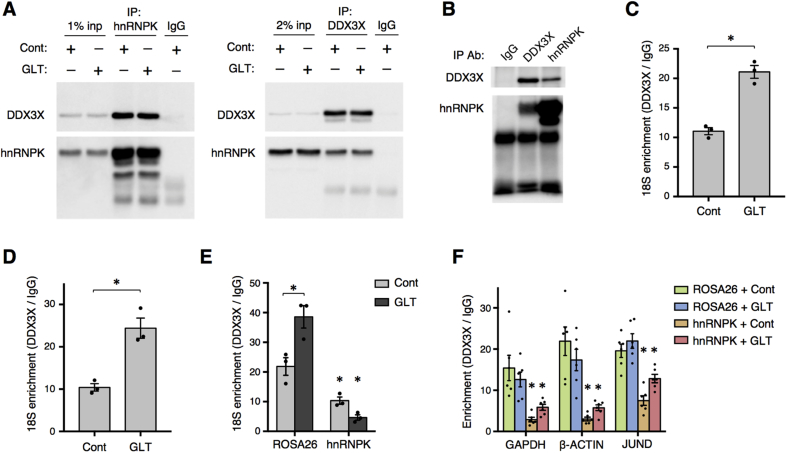
Figure 6.
hnRNPK interacts with DDX3X and promotes DDX3X-18S interaction. (A) Western blots from pull-down of hnRNPK (left) or DDX3X (right) in Min6 cells treated with control (Cont) or glucolipotoxic (GLT) conditions for 30 h. (B) Representative Western blot from pull-down of hnRNPK and DDX3X in mouse islets indicating interaction between these factors in primary tissue. (C) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in Min6 cells treated with Cont or GLT conditions for 30hrs (n = 3). (D) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in mouse islets treated with Cont or GLT conditions for 2 days (n = 3). (E) RNA immunoprecipitation for DDX3X followed by RT-qPCR for 18S ribosomal RNA in Min6 cells with CRISPR-mediated depletion of hnRNPK and cultured in Cont or GLT conditions for 30hrs (n = 3). (F) Decreased interaction between DDX3X and indicated mRNAs after CRISPR-mediated depletion of hnRNPK in Min6 cells treated with Cont or GLT conditions for 30hrs as determined by RNA immunoprecipitation for DDX3X (n = 3). P values were calculated by two-way ANOVA except in (C) and (D) in which unpaired two-tailed Student's t-tests were used. In (E,F), * denotes significance compared to ROSA26 group of the same treatment unless otherwise noted. * = p < 0.05. Data are presented as mean ± standard error of the mean unless otherwise noted.