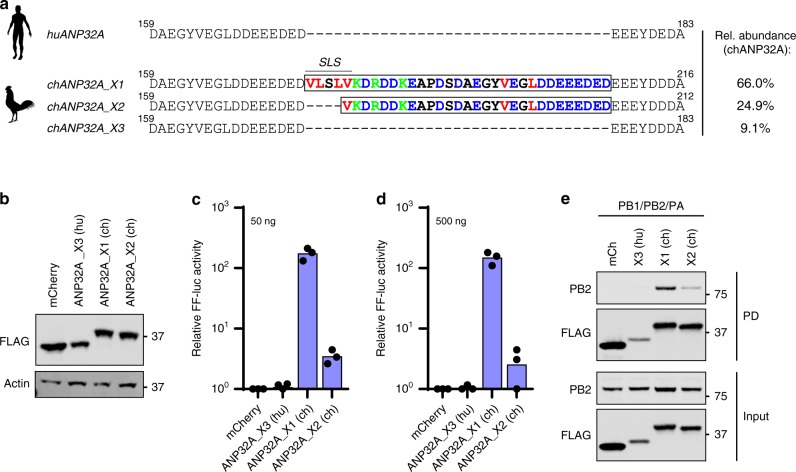
Fig. 1.
Avian species express at least three ANP32A variants that differentially impact avian-signature IAV polymerase activity. a Sequence alignment between human ANP32A (huANP32A, Homo sapiens, NP_006296.1) and chicken ANP32A (chANP32A, Gallus gallus_X1, XP_413932.3; Gallus gallus_X2, XP_004943985.1; Gallus gallus_X3, XP_025009881.1). Hydrophobic residues (red), acidic stretches (blue) and basic residues (green), are highlighted. SLS, SIM-like sequence. Relative abundance of ANP32A isoform transcripts in chicken DF-1 cells, as determined experimentally by NGS quantification of cDNA-derived amplicons, is shown on the right. b Western blot analysis of lysates from human 293T cells transfected with the indicated FLAG-ANP32A constructs. c Polymerase reconstitution assay comparing the impact of each FLAG-ANP32A construct (50 ng) on PB2-627E vPol activity in human 293T cells. d Similar to c, but using 500 ng of each FLAG-ANP32A construct. In panels c, d, bars represent mean values from three independent experiments, with the individual data points shown. e 293T cells were transfected with the indicated FLAG-tagged constructs together with PB1, PA, and PB2 (627E). Following anti-FLAG precipitation (PD), the indicated proteins were detected by western blot. For panels b and e, representative data from two independent experiments are shown. Source data for panels b–e are provided in the Source Data file