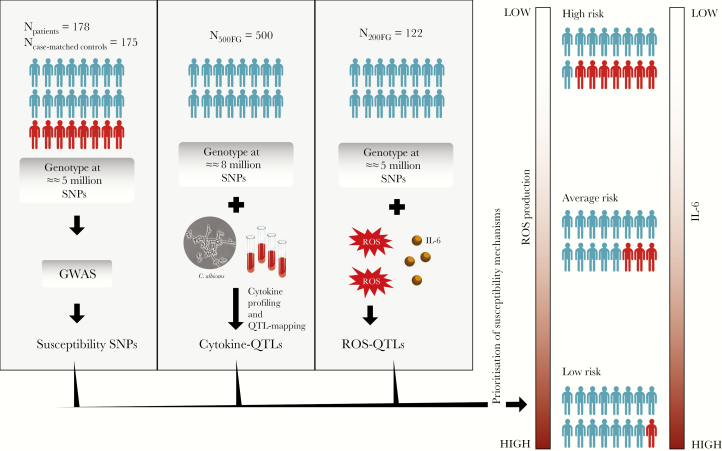
Figure 1.
Study overview. We performed a genome-wide association study (GWAS) to test the association of genetic variants to candidemia susceptibility by using 178 patients and 175 case-matched controls. To identify whether susceptibility variants influence cytokine levels in response to Candida albicans, we first collected blood samples from 500 healthy individuals (500FG cohort), isolated their DNA, and genotyped it. Whole blood or peripheral blood mononuclear cells (PBMCs) or monocyte-derived macrophages were also isolated to perform a series of stimulation experiments with C albicans to profile cytokines released in the 3 cell systems. Using the genotype information and cytokine measurements, we mapped Candida-response cytokine quantitative trait loci (QTLs) in the 500FG cohort. Finally, we generated genotype data and measured reactive oxygen species (ROS) production from Candida-stimulated PBMCs isolated from an independent cohort of 122 healthy volunteers (200FG cohort). In addition, we profiled cytokine interleukin (IL)-6 produced from PBMCs isolated from a subgroup of 200FG cohort (n = 61). Taken all datasets, we showed that the numbers of risk alleles at candidemia loci, which influence cytokine levels in response to Candida, are negatively correlated with ROS production in response to Candida. Finally, ROS production showed a positive correlation with IL-6 measurements in 200FG. SNPs, single-nucleotide polymorphisms.