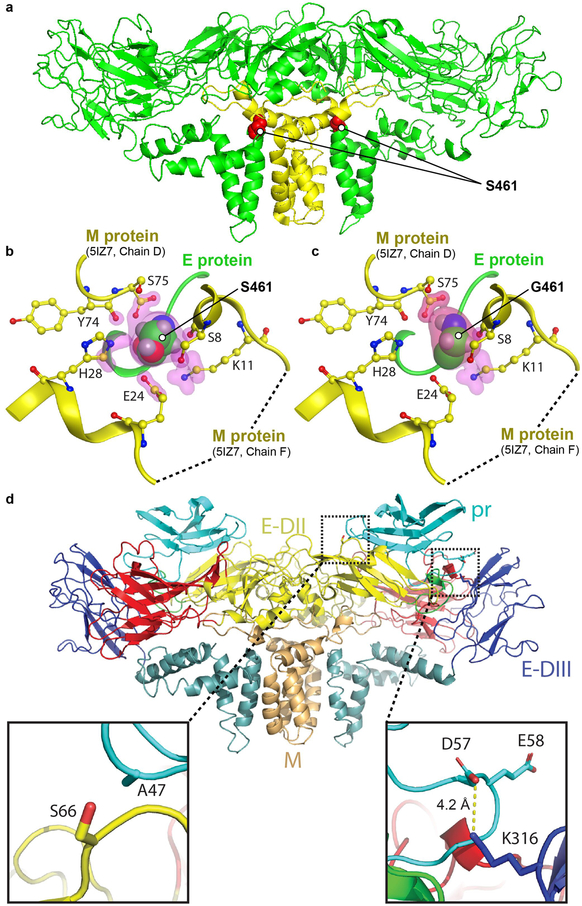
Figure 4.
Molecular analysis and modelling of the S461G and K316 mutations. a) Cryo-EM structure of H/PF/3013 strain ZIKV E dimer (PDB accession number: 5IZ7) showing the position of the S461G mutation (red spheres) in the mature structure. E and M proteins represented by green and yellow ribbon structures, respectively. b) Residue S461 sits in a pocket -in contact with six M protein residues; Y74, S75 (from chain D in M protein) and S8, K11, E24, H28 (from chain F in M protein). Contact atoms are surrounded by pink highlights. Protein contact analysis by Molecular Operating Environment (MOE) software package (using 5IZ7). c) The S461G substitution results in only 3 residues in contact with G461. The S461G mutation was introduced on the 5IZ7 structure followed by energy minimization and protein contact analysis by MOE. d) Homology model of ZIKV Natal pr protein (based on DENV structure PDB:3C5X and generated using Swiss-Model) fitted to the cryo-EM structure for low pH immature DENV (EMD-5006), showing a potential salt bridge forming between D57 in pr and K316 in E-DIII (right inset). A S66L mutation can potentially result in hydrophobic interaction between L66 in E-DII and A47 in pr (left inset).