Abstract
Objectives
We investigated the genetic history of southern African populations with a special focus on their paternal history. We reexamined previous claims that the Y‐chromosome haplogroup E1b1b (E‐M293) was brought to southern Africa by pastoralists from eastern Africa, and investigated patterns of sex‐biased gene flow in southern Africa.
Materials and methods
We analyzed previously published complete mtDNA genome sequences and ∼900 kb of NRY sequences from 23 populations from Namibia, Botswana, and Zambia, as well as haplogroup frequencies from a large sample of southern African populations and 23 newly genotyped Y‐linked STR loci for samples assigned to haplogroup E1b1b.
Results
Our results support an eastern African origin for Y‐chromosome haplogroup E1b1b (E‐M293); however, its current distribution in southern Africa is not strongly associated with pastoralism, suggesting more complex demographic events and/or changes in subsistence practices in this region. The Bantu expansion in southern Africa had a notable genetic impact and was probably a rapid, male‐dominated expansion. Our finding of a significant increase in the intensity of the sex‐biased gene flow from north to south may reflect changes in the social dynamics between Khoisan and Bantu groups over time.
Conclusions
Our study shows that the population history of southern Africa has been complex, with different immigrating groups mixing to different degrees with the autochthonous populations. The Bantu expansion led to heavily sex‐biased admixture as a result of interactions between Khoisan females and Bantu males, with a geographic gradient which may reflect changes in the social dynamics between Khoisan and Bantu groups over time.
Keywords: admixture, Bantu, Khoisan, mtDNA, NRY
1. INTRODUCTION
The extensive genetic, linguistic, and cultural diversity of southern African populations (Barbieri et al., 2014, 2016; Barnard, 1992; Güldemann, 2008, 2014; Marks et al., 2015; Oliveira et al., 2018; Pickrell et al., 2012; Salas et al., 2002; Schlebusch et al., 2011; Tishkoff et al., 2007; Wood et al., 2005) reflects a long history of population movements and interactions. The so‐called Khoisan populations are the descendants of some of the earliest humans inhabiting the region; they are or used to be foragers and pastoralists who speak indigenous non‐Bantu languages characterized by the heavy use of click consonants. We use the term “Khoisan” without any assumption about their genetic or linguistic unity (cf. Barnard, 1992). Three language families are recognized among Khoisan (Supporting Information Figure S1): Tuu and Kx'a, spoken by populations known to have practiced hunting and gathering until recently, and Khoe‐Kwadi, spoken by a large number of different ethnolinguistic groups practicing diverse subsistence strategies (Güldemann, 2004, 2005, 2008, 2014; Güldemann & Elderkin, 2010; Heine & Honken, 2010). Genetic data revealed that the Khoisan populations harbor some of the earliest branching mtDNA and NRY lineages (Barbieri et al., 2013; Barbieri et al., 2014, 2016; Rosa & Brehm, 2011; Tishkoff et al., 2007). Additionally, autosomal genetic data indicate complex patterns of ancestry for most Khoisan groups, reflecting substantial admixture with other groups as well as between different Khoisan groups (Montinaro et al., 2017; Pickrell et al., 2012; Pickrell et al., 2014; Uren et al., 2016).
It has been shown that there are at least two different sets of related episodes of gene flow in the history of Khoisan populations that could have contributed to their current genetic ancestry. The earlier admixture event involves a migration from eastern Africa that occurred 900–1,800 years ago (Montinaro et al., 2017; Pickrell et al., 2014; Schlebusch et al., 2017) and is supported by several independent lines of evidence from different disciplines. Archeological data support an introduction of pastoralism from eastern to southern Africa (Mitchell, 2002; Pleurdeau et al., 2012), while based on linguistic data it has been hypothesized that Khoe‐Kwadi‐speaking populations are the descendants of these pastoralist migrants from eastern Africa (Güldemann, 2008), where livestock is present from 4,000 years ago (Deacon & Deacon, 1999; Phillipson, 2005). Genetic evidence of shared ancestry between the Khoe‐Kwadi‐speaking Nama pastoralists and the ǂKhomani and Karretjie (whose heritage languages belonged at least in part to the Tuu family), and East African groups, specifically the Maasai, was observed in autosomal data (Schlebusch et al., 2012). Recent studies of ancient DNA from skeletal remains from Africa demonstrated that all modern‐day Khoisan groups for which there are genetic data have been influenced by 9%–22% genetic admixture from East African/Eurasian pastoralist groups (Schlebusch et al., 2017; Skoglund et al., 2017).
Further evidence for a migration from East Africa comes from elevated frequencies of an East African lactase persistence allele in southern African pastoralist groups and in Khoe‐speaking groups, particularly the Nama (Breton et al., 2014; Macholdt et al., 2014; Macholdt, Slatkin, Pakendorf, & Stoneking, 2015; Schlebusch et al., 2012), and mtDNA haplogroup L4b2. This haplogroup is found in southern African Nama and ǂKhomani San, as well as in high frequency in the Hadza and Sandawe from Tanzania (Knight et al., 2003; Tishkoff et al., 2007), who also make use of click consonants in their languages.
Additional support for a demic diffusion from East Africa is based on the distribution of Y‐chromosome haplogroup E1b1b (E‐M293). It has been suggested that this haplogroup spread through Tanzania to southern‐central Africa via a movement of people who brought pastoralism ∼2,000 years ago (Henn et al., 2008), independently of the migration of Bantu‐speaking peoples. New studies of autosomal data suggest that pastoralism, after being brought to southern Africa from eastern Africa via at least some degree of demic migration, was subsequently spread within southern Africa mostly via cultural diffusion (Montinaro et al., 2017; Uren et al., 2016).
The more recent admixture event reconstructed with genomic data is a consequence of the Bantu expansion that started around 5,000 years ago from the current Cameroon‐Nigeria border. This expansion is one of the most influential demographic events on the African continent (Grollemund et al., 2015), and led to a sex‐biased pattern of admixture between Bantu speakers and the local groups already present in territories settled by Bantu‐speaking groups, including forager populations such as the rain forest hunter‐gatherers in Central Africa and the Khoisan groups of southern Africa (Destro‐Bisol et al., 2004; Schlebusch et al., 2011; Tishkoff et al., 2007; Verdu et al., 2009, 2013; Wood et al., 2005). This sex‐biased pattern is the result of mating practices that typically involve Bantu males and autochthonous females, but hardly ever involve autochthonous males and Bantu females (Destro‐Bisol et al., 2004). This results in the flow of Bantu Y‐chromosomes into autochthonous communities, or of autochthonous mtDNAs into Bantu communities, depending on where the children are raised.
In this study, we use previously published mtDNA and NRY sequences collected from a large and comprehensive sample of Khoisan and Bantu‐speaking populations to investigate (1) the genetic history and structure of southern African populations, with a focus on their previously undescribed paternal genetic history as well as the distribution of haplogroups and specific lineages within different ethnolinguistic groups; (2) the link between the NRY haplogroup E1b1b (E‐M293) and pastoralism; and (3) the intensity of the sex‐biased gene flow between incoming non‐autochthonous and autochthonous peoples.
2. MATERIALS AND METHODS
2.1. Samples
We collected data from two available datasets which analyzed the same population samples. The Y‐chromosome dataset consists of ∼900 kb sequences from the non‐recombining region from 547 individuals belonging to 24 different populations (Supporting Information Table S1 and Figure 1; Barbieri et al., 2016), and the mtDNA dataset comprises complete mtDNA genome sequences from 680 individuals belonging to 26 different populations (Barbieri et al., 2014). The NRY sequences from the neighboring Khwe‐speaking ǁAni and Buga populations were merged together into a combined ǁAni/Buga population due to their low sample sizes; the mtDNA sequences were similarly merged to be directly comparable to the NRY dataset. We refer to the dataset of 23 populations (17 Khoisan and six Bantu) that overlap at the level of populations between the NRY and mtDNA datasets from Namibia, Botswana and Zambia, as the “NBZ dataset” (Supporting Information Table S2). Approximately 93% of the individuals included in the NRY dataset are also included in the mtDNA dataset, while ∼73% of the individuals included in the mtDNA dataset are also included in the NRY dataset. For analyses of autochthonous genetic structure before non‐autochthonous haplogroups arrived in the area, we excluded from the NBZ data sets (both mtDNA and the NRY) all Bantu populations, non‐autochthonous haplogroups from Khoisan populations, and Khoisan populations with sample sizes <8 after removal of individuals with non‐autochthonous haplogroups. This filtering resulted in 13 populations overlapping between mtDNA and the NRY, and we refer to this dataset as the “AU‐NBZ dataset” (Supporting Information Table S2). The analysis of the intensity of the sex‐biased gene flow (ISBGF) included additional data from southern African populations for which both mtDNA and NRY haplogroup frequencies were previously published (Barbieri, Butthof, Bostoen, & Pakendorf, 2013; Coelho, Sequeira, Luiselli, Beleza, & Rocha, 2009; de Filippo, Heyn, Barham, Stoneking, & Pakendorf, 2010; Henn et al., 2011; Marks et al., 2015; Schlebusch et al., 2011), and we refer to this as the “SA dataset” (Supporting Information Table S3). The highly admixed Karretjie and Colesberg Coloured populations from South Africa were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (see Schlebusch et al., 2011; Traill, 1996).
Figure 1.
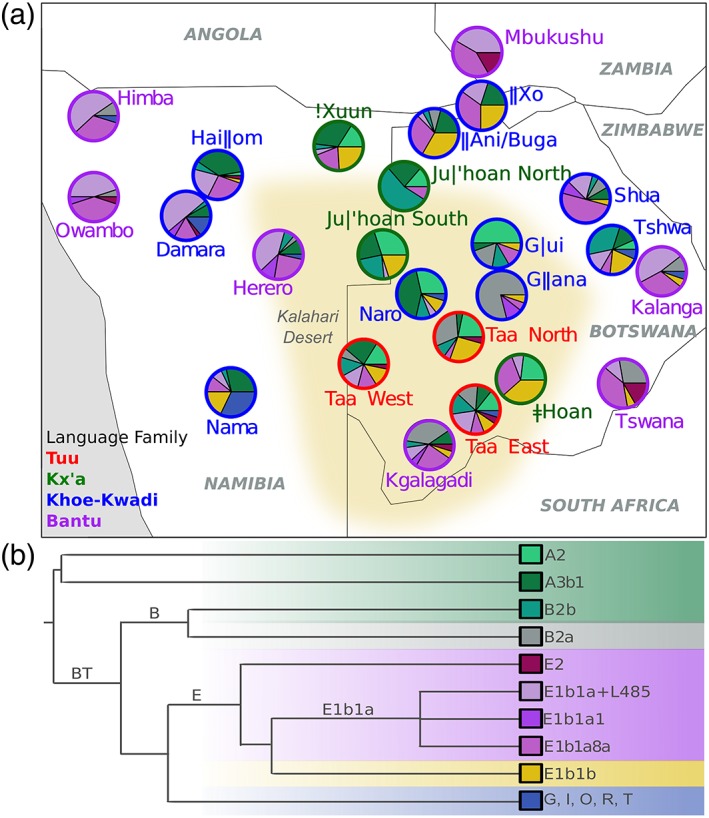
(a) Map of the approximate location of the populations and their NRY haplogroup composition. Population labels are color‐coded according to linguistic affiliation as indicated in the lower left corner of the map. Green shades within the pie charts indicate haplogroups that are traditionally considered as Khoisan‐related, purple shades indicate Bantu‐related haplogroups, yellow indicates E1b1b (thought to be East African), blue are Eurasian haplogroups, and gray represents B2a, which is probably associated with both Khoisan and Bantu populations. The Bantu‐related, Eurasian, and E1b1b haplogroups are here defined as non‐autochthonous haplogroups, while Khoisan‐related haplogroups are defined as autochthonous. The yellow area indicates the Kalahari Desert. (b) Schematic representation of the NRY phylogenetic tree, from Barbieri et al. (2016) [Color figure can be viewed at wileyonlinelibrary.com]
Since the time of sample collection, additional linguistic research on the Kx'a family has revealed that the language formerly referred to as ǂHoan consists of three dialects: N!aqriaxe, ǂHoan, and Sasi. The language is nowadays referred to as ǂ'Amkoe (Gerlach, 2016; Güldemann, 2014). Although the samples included under the name ǂHoan mainly stem from N!aqriaxe speakers and include only a few ǂHoan speakers, for ease of comparison with previous studies of these samples we continue to refer to them as ǂHoan speakers.
Most of the individuals assigned to major haplogroup E1b1b (n = 59) were assigned to E1b1b1b2b (E‐M293; n = 36), while due to lower coverage the rest were assigned to E1b1b1b2 (E‐CTS5487, n = 23). Manual inspection confirmed that the individuals carrying the derived allele for M293 are randomly distributed in all sub‐branches within the E1b1b sequence‐based network, and thus we assumed that even those individuals where the position was not successfully genotyped are derived for marker E‐M293 (the marker genotyped by Henn et al., 2008). Individuals assigned to E1b1b were genotyped for a set of 23 STRs using the PowerPlex® Y23 System (Promega, Mannheim, Germany; Supporting Information Table S4A) as described previously (Barbieri et al., 2016). To place the southern African samples in a broader picture and search for possible connections with eastern Africa, they were subsequently merged with publicly available STR datasets for the E1b1b haplogroup from Africa (Berniell‐Lee et al., 2009; de Filippo et al., 2011; Gomes, Sánchez‐Diz, Amorim, Carracedo, & Gusmão, 2010; Henn et al., 2008; Tishkoff et al., 2007), resulting in the “E1b1b‐STR dataset” (Supporting Information Table S4B) with a total of 278 individuals with 10 overlapping STRs (DYS19, DYS389I, DYS389II, DYS390, DYS391, DYS392, DYS393, DYS437, DYS438, and DYS439).
2.2. Data analysis
We used previous haplogroup assignments for both the mtDNA and the NRY data (Barbieri et al., 2014, 2016; Barbieri, Vicente, et al., 2013). A network for the NRY sequences was generated previously by Barbieri et al. (2016), but was here analyzed by population for the first time to investigate the distribution of specific lineages within different ethnolinguistic groups. Branches in the NRY network were numbered according to the tree in Figure 1 of Barbieri et al. (2016). Additionally, Network 5.0.0.1. (Fluxus Engineering, http://www.fluxus-engineering.com) was used to visualize the relationships between the STR haplotypes genotyped for haplogroup E1b1b and previously published for B2a (Barbieri et al., 2016). We excluded DYS385 a/b, as the PCR analysis co‐amplifies these two loci (so alleles cannot be definitively assigned to a locus). The STR‐based networks were constructed by either applying the reduced‐median method first and then the median‐joining method (network with 10 STRs; Bandelt, Forster, & Röhl, 1999), or just the median‐joining method (networks with 21 STRs) with STRs weighted according to their mutation rate (Heinila, 2012). Networks were subsequently plotted and colored with Network Publisher.
Analyses of Molecular Variance (AMOVA; Excoffier, Smouse, & Quattro, 1992) and matrices of pairwise Φ ST distances were computed in Arlequin ver. 3.11 (Excoffier, Laval, & Schneider, 2005) for both the NBZ and AU‐NBZ dataset. For the NRY calculations, we used a Tamura and Nei (TrN) model (Tamura & Nei, 1993) with a gamma value of 155.8, and for the mtDNA, a TrN model with a gamma value of 0.26, as suggested by results obtained with jModelTest 2 (Darriba, Taboada, Doallo, & Posada, 2012). An AMOVA was performed in parallel for both the mtDNA and NRY to investigate previously proposed groupings and to explore possible factors that might influence genetic structure. Detailed information about the AMOVA groupings and populations included in each of the groups can be found in Supporting Information Table S5. Ecotype information was obtained from the WWF Terrestrial Ecoregions of the World (TEOW) map (Olson et al., 2001) and populations were grouped accordingly. Nonmetric multidimensional scaling (MDS) analyses were performed with the function “isoMDS” from the package MASS (Venables & Ripley, 2002). Neighbor‐joining trees (NJ trees) depicting population relationships were generated from the matrix of Φ ST distances with the function “nj” of the package ape (Paradis, Claude, & Strimmer, 2004). Correspondence analysis (CA) was performed with the package ca (Nenadić & Greenacre, 2007), using haplogroup frequencies as input variables. A Mantel test was performed between genetic (Φ ST) and geographic distances with the R package ade4 (Chessel, Dufour, & Thioulouse, 2004); geographic distances between populations were averaged over GPS data from the individual sampling locations with the function rdist.earth of the package fields (Nychka et al., 2017).
The intensity of the sex‐biased gene flow (ISBGF) was calculated for the SA dataset as the difference between the proportion of autochthonous mtDNA haplogroups (L0d, L0k) and autochthonous NRY haplogroups (A2, A3b1, B2b) for each of the populations after removing Eurasian haplogroups in order to avoid recent sex‐biased gene flow from European colonists (i.e., NRY haplogroups G, I, O, R, T): ISBGF = %(L0d + L0k) − %(A2 + A3b1 + B2b). We calculated the standard deviation of ISBGF by performing a bootstrap analysis with 100 replicates for each population (see Supporting Information Table S3). For this, we sampled mtDNA haplogroups and Y‐chromosome haplogroups that were present in the population independently with replacement, calculated for each marker the frequency of autochthonous haplogroups and subsequently the ISBGF. Population sizes were kept constant throughout the bootstrap analysis. It is not clear if NRY haplogroup B2a is autochthonous to southern Africa or was brought by the Bantu expansion (or possibly both; see Barbieri et al., 2016); we therefore treated haplogroup B2a as ambiguous and ran analyses in parallel treating it as either autochthonous (AUT) or non‐autochthonous (NAUT). Values close to zero indicate equal proportions of autochthonous uniparental markers in a given population; positive values indicate a higher proportion of autochthonous mtDNA haplogroups than autochthonous NRY haplogroups; negative values indicate a higher proportion of autochthonous NRY haplogroups than autochthonous mtDNA haplogroups. The p values for the AMOVA, Mantel Test, and correlations between ISBGF and longitude and latitude were corrected for multiple testing with the “fdr” method using the function p.adjust from the R package stats (R Core Team, 2014).
3. RESULTS
3.1. Y‐chromosome lineages in southern Africa
3.1.1. Khoisan‐related haplogroups
Traditionally, the A2 (A‐V50), A3b1 (A‐M51) and B2b (B‐M192) Y‐chromosome lineages are described as haplogroups characteristic of the autochthonous southern African Khoisan populations of foragers and pastoralists (Barbieri et al., 2016; Batini et al., 2011; Marks et al., 2015; Underhill et al., 2000; Wood et al., 2005). Even though macro‐haplogroups A2 and B2b are also found in Central African rain forest hunter‐gatherers, different subclades are characteristic of Khoisan populations of southern Africa (Batini et al., 2011). In our dataset Khoisan‐related haplogroups are, as expected, observed in higher frequencies in Khoisan than in Bantu populations (Supporting Information Figure S2). However, these haplogroups are also observed in relatively high frequency (>14%) in two Bantu‐speaking populations (Herero and Kgalagadi; Supporting Information Table S6).
The A2 (A‐V50) haplogroup has a narrow distribution in the central Kalahari (Figure 1 and Supporting Information Table S6), and it has been detected in low frequency in Baka foragers from Cameroon and Gabon (Batini et al., 2011). In the network of A2 sequences (Supporting Information Figure S3), all but one haplotype of the Tuu‐speaking populations occur on branch 9 (see Table S1 of Barbieri et al., 2016 for more information about branches), while Kx'a and Khoe‐Kwadi populations are present in most of the other branches. All !Xuun, Juǀ'hoan South and Juǀ'hoan North haplotypes occur in the same branches as Naro haplotypes (for reasons of simplicity from here on, we refer to Juǀ'hoan North and Juǀ'hoan South together as Juǀ'hoan; and to Taa West, Taa North, and Taa East together as simply Taa). The ǂHoan haplotypes are more closely related to haplotypes from neighboring Taa and Gǀui populations than to haplotypes from other Kx'a speakers.
Haplogroup A3b1 (A‐M51) shows strong regional and linguistic clustering. It represents a major autochthonous NRY haplogroup in most Khoe‐Kwadi‐speaking populations (ranging in frequency from 0% to 43%; Supporting Information Table S6), and is the only autochthonous haplogroup in the Nama. Interestingly, almost all haplotypes from Khoekhoe speakers (Nama, Haiǁom and Damara) and !Xuun are found in a single branch of the network (branch 13; Supporting Information Figure S4). Another branch (branch 11; Supporting Information Figure S4) almost exclusively contains haplotypes found in the Khoe‐Kwadi‐speaking Khwe (ǁXo and ǁAni/Buga) and Naro, which inhabit the Okavango delta and neighboring Ghanzi District, respectively. The Juǀ'hoan and Taa populations make up the majority of the haplotypes found in branch 18, and they harbor similar yet distinct sublineages within this branch.
Present in almost all Khoisan populations (except ǁXo, Gǁana and ǂHoan), B2b (B‐M192) has frequencies higher than 23% in Juǀ'hoan and Tshwa, and frequencies lower than 15% in the other populations (Supporting Information Table S6). All but two haplotypes from the Kx'a‐speaking Juǀ'hoan and !Xuun belong to branch 26 in the network (Supporting Information Figure S5), which they share with all Naro haplotypes and one Haiǁom haplotype. All Taa haplotypes are found in two distinct sublineages within branch 26 (Supporting Information Figure S5).
3.1.2. B2a (B‐M150) haplogroup
Although the B2a haplogroup was previously treated as an indicator of Bantu gene flow (Batini et al., 2011; Beleza, Gusmão, Amorim, Carracedo, & Salas, 2005; Berniell‐Lee et al., 2009; Quintana‐Murci et al., 2010), Barbieri et al. (2016) showed that this haplogroup might have existed in Khoisan populations before the arrival of Bantu speakers. Given its ambiguous origin, we ran analyses in parallel treating it as either autochthonous or non‐autochthonous. The highest frequency of this haplogroup (∼80%) is found in the Gǁana (Figure 1 and Supporting Information Table S6). In addition, B2a is also found in three other Khoe‐Kwadi populations (ǁAni/Buga, Gǀui, and Shua), all Tuu‐speaking populations, and all Bantu populations except Mbukushu. It is absent from all Kx'a‐speaking populations and the remaining Khoe‐Kwadi populations. All of the haplotypes from the West Bantu‐speaking Himba, Herero, and Owambo are in a specific sublineage separate from other haplotypes (dotted circle in Supporting Information Figure S6a), while the East Bantu‐speaking Kalanga, Tswana and Kgalagadi haplotypes are found in a star‐like cluster together with haplotypes from Khoisan populations (mostly from central Kalahari Taa and Gǁana populations).
3.1.3. Bantu‐related haplogroups
Bantu‐associated haplogroups such as E1b1a (E‐M2) and E2 (E‐M75) (Barbieri et al., 2016; de Filippo et al., 2011; Quintana‐Murci et al., 2010) are found at frequencies higher than 66% in all Bantu populations except Kgalagadi (43%), while in Khoisan populations their frequency ranges between 3 and 75%, with an average of 29% (Figure 1 and Supporting Information Figure S2 and Table S6). The most striking pattern is that most of the Tuu and Kx'a‐speaking groups have low proportions of Bantu‐related haplogroups (range 3%–38%), while Khoe‐Kwadi‐speaking groups vary much more (range 5%–75%).
The network for haplogroup E1b1a + L485 (E‐L485) sequences (Supporting Information Figure S7) shows a star‐like pattern, suggestive of population expansion, that harbors haplotypes from all language families. The Damara haplotypes are found within branch 38, and they are similar to the haplotypes found in neighboring West Bantu‐speaking Himba, Herero and Owambo. Interestingly, a newly described sub‐lineage (Barbieri et al., 2016) of this haplogroup (branch 35) is present exclusively in Khoe‐Kwadi‐speaking groups, namely Khwe (ǁAni/Buga and ǁXo) and Haiǁom. The similarity of Damara and West Bantu haplotypes is also noticeable in branch 39 of the haplogroup E1b1a1 (E‐M58) network (Supporting Information Figure S7). The network for haplogroup E1b1a8a (E‐U175) sequences is similar to that of haplogroup E1b1a + L485 in exhibiting a star‐like pattern with haplotypes from all of the language families in the core (Supporting Information Figure S8). Finally, haplogroup E2 (E‐M75) is found in frequencies lower than 5% in Taa, Damara, Haiǁom, Owambo, and Kgalagadi, while in Mbukushu and Tswana it reaches almost 17% (Figure 1 and Supporting Information Table S6 and Figure S9).
3.1.4. E1b1b (E‐M293) haplogroup
This haplogroup is considered to have an East African origin, and it has been associated with the spread of pastoralism from East Africa to southern Africa (Henn et al., 2008; Trombetta et al., 2015). The sequence‐based network of this haplogroup shows a star‐like pattern with all language families represented in the core of the network (Supporting Information Figure S10A). Interestingly, most of the Khoekhoe‐speaking Nama haplotypes are found in the core. Haplotypes found in the ǂHoan individuals are on a branch shared with Taa and Gǀui haplotypes (dotted circle in Supporting Information Figure S10A). The analysis of this haplogroup based on STR data and its possible link to the spread of pastoralism is discussed in detail below.
3.1.5. Eurasian‐related haplogroups (CF‐P143)
In eight populations, we found 20 individuals (Supporting Information Table S1) with NRY haplogroups that are traditionally considered to be of Eurasian origin (Underhill & Kivisild, 2007).
3.1.6. AMOVA
Factors that might influence the genetic structure of southern African populations can be explored by grouping populations using various criteria and then examining the proportion of genetic variance shared among groups, among populations within groups, and within populations, using AMOVA (Excoffier et al., 1992). When all Khoisan populations are analyzed together as one group in the AMOVA, they show slightly higher differentiation between populations for mtDNA than for the NRY (∼17% vs ∼15%; Table 1), which may reflect geographically structured mtDNA lineages and recent expansion of Bantu‐related NRY haplogroups. However, this pattern varies when the Khoisan language families are analyzed separately; the Khoe‐Kwadi harbor the biggest proportion of between‐population variance for both uniparental markers. Overall, Bantu groups harbor levels of between‐group variation that are comparable to Khoisan populations for mtDNA but lower for the NRY (Table 1), in keeping with other evidence that the Bantu speakers incorporated more female than male lineages from autochthonous populations, and that Khoisan populations incorporated more male than female Bantu lineages (Destro‐Bisol et al., 2004; Marks et al., 2015; Pakendorf, Bostoen, & de Filippo, 2011; Schlebusch et al., 2011; Tishkoff et al., 2007; Verdu et al., 2013; Wood et al., 2005).
Table 1.
Analysis of molecular variance (AMOVA) for the mtDNA and NRY data
| mtDNA | NRY | |||||
|---|---|---|---|---|---|---|
| Among groups | Among populations within groups | Within populations | NBZ AMOVA grouping | Among groups | Among populations within groups | Within populations |
| 21.26** | 78.74 | All populations (1) | 17.08** | 82.92 | ||
| 17.27** | 82.73 | Khoisan (1) | 14.69** | 85.31 | ||
| 18.18** | 81.82 | Khoe‐Kwadi (1) | 18.79** | 81.21 | ||
| 7.35** | 92.65 | Kx'a (1) | 10.16** | 89.84 | ||
| 8.12** | 91.88 | Tuu (1) | 1.17 | 98.83 | ||
| 16.61** | 83.39 | Bantu (1) | 8.29** | 91.71 | ||
| 18.46** | 5.37** | 76.17 | Uren (5) | 12.6** | 6.33** | 81.07 |
| 1.57 | 19.91** | 78.52 | WWF (7) | 1.24 | 16.01** | 82.75 |
| 14.61** | 10.46** | 74.93 | Subsistence (4) | 8.15** | 10.96** | 80.89 |
| 18.86** | 3.9** | 77.24 | Geo‐linguistic (8) | 9.4** | 8.49** | 82.11 |
| 13.89** | 14.75** | 71.36 | Linguistics (2) | 9.46** | 12.57** | 77.97 |
| 8.77** | 14.49** | 76.74 | Linguistics (4) | 5.84* | 12.58** | 81.58 |
The names of different groupings are followed by the number of groups defined in brackets (see Supporting Information Table S5 for information on populations included in each of the groups). FDR‐corrected p values significant at the 0.05 and 0.01 levels are indicated with * and **, respectively.
The highest between‐group variance for the NRY, and one of the highest for mtDNA, is seen when populations are grouped into five groups defined by distinct, geographically organized autosomal ancestry components inferred from unsupervised population structure analysis (Uren et al., 2016). Based on the distribution of autosomal ancestries they defined, Uren et al. (2016) concluded that the autosomal structure in Khoisan populations reflects the role of geographic barriers and the ecology of the greater Kalahari Basin. In order to test if our dataset of uniparental markers is in agreement with this conclusion, we obtained ecotype information from the WWF Terrestrial Ecoregions of the World (TEOW) map (Olson et al., 2001) and grouped populations accordingly (WWF, Table 1). The much lower among‐group variance for this grouping (for both uniparental markers) suggests that the ecoregions in which the populations currently live does not explain the genetic structure of the studied populations. Grouping populations by the geo‐linguistic categories defined by Barbieri et al. (2014) results in capturing ∼19% of the variance seen in mtDNA between eight geo‐linguistic categories (Geo‐linguistic, Table 1) and is thus one of the best groupings for the mtDNA, but it captures just ∼9.4% variation for the NRY. When only linguistic criteria are applied, grouping by the two major linguistic groups (Bantu and Khoisan) better explains the variance between groups for both mtDNA and the NRY than when grouping by the four language families (Bantu, Kx'a, Tuu, and Khoe‐Kwadi).
The results of the AMOVA analysis for the AU‐NBZ data set, that is, after removing the Bantu populations and non‐autochthonous uniparental lineages (Supporting Information Table S7) reveals that there is larger genetic differentiation among groups for the NRY than for the mtDNA (∼23% vs ∼8%), in contrast to the full dataset (∼17% vs ∼21%), suggesting differences in male vs. female migration between Khoisan and Bantu groups, a hypothesis that is analyzed in more detail below. However, although the AU‐NBZ dataset was constructed to try to investigate Khoisan population structure prior to the arrival of Bantu populations, the surviving autochthonous genetic structure was undoubtedly influenced by contact with Bantu populations, that is, probably many autochthonous lineages were subsequently lost.
3.1.7. ΦST distance‐based analysis: MDS and NJ tree
To further investigate the differentiation and relationship among populations, we performed MDS analysis and constructed NJ trees. Bantu‐speaking populations are separated from Kx'a and Tuu speakers in both the mtDNA and the NRY MDS (Figure 2), while the Khoe‐Kwadi populations are spread between them. As expected, this pattern is also noticeable in the mtDNA and the NRY NJ trees (Supporting Information Figure S11a,b), where Bantu populations tend to be on the opposite side of the tree compared to Kx'a and Tuu‐speaking populations. The Damara appear to be closer to Bantu‐speaking populations than to the majority of the Khoisan populations for both mtDNA and the NRY, while the Gǁana population is a clear outlier in the NRY MDS, with the Kgalagadi and Taa North as the closest populations. Within the Bantu populations, Kgalagadi and Tswana from the central Kalahari are more distant from the rest of the Bantu populations for both mtDNA and the NRY (Figure 2).
Figure 2.
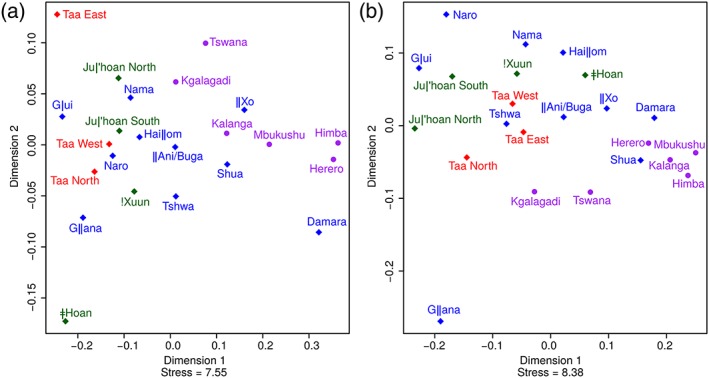
Multidimensional scaling plot based on (a) mtDNA and (b) NRY ΦST distances (Supporting Information, Table S11). Population symbols and colors indicate linguistic affiliation, as shown in Figure 1 [Color figure can be viewed at wileyonlinelibrary.com]
3.1.8. Correspondence analysis (CA)
Considering that Bantu and Khoisan populations are differentiated at the level of uniparental haplogroups, we did CA to visualize the relationships among them based on haplogroup frequencies. In contrast to the Φ ST distance‐based analyses, the distinction between Khoisan and Bantu populations in the CA (Supporting Information Figure S11c,d) is striking. Most Khoisan populations exhibit low differentiation for mtDNA haplogroups due to high frequencies of the autochthonous haplogroups L0d and L0k, whereas in the NRY CA plot most of the Bantu populations are clustered together due to their high frequency of Bantu‐related NRY haplogroups, such as E1b1a and E2. The Khoe‐Kwadi populations are dispersed between Bantu populations on the one side and Kx'a and Tuu populations on the other side. Although the Gǁana group with other Khoisan populations in the mtDNA CA plot, they are a clear outlier in the NRY CA plot, due to their high frequency of haplogroup B2a. As in the MDS analysis, the mtDNA and NRY CA plots show the Damara closer to the Bantu‐speaking pastoralist Himba and Herero populations than to other Khoisan populations.
3.1.9. Mantel Test: geographic vs genetic distances
To asses if geographically proximate populations are genetically closer to each other, we performed a Mantel Test. There is a statistically significant correlation between pairwise geographic distances and mtDNA Φ ST distances (Mantel Test, R 2 = 0.258, FDR‐corrected p value = 0.02), but not between pairwise geographic distances and NRY Φ ST distances (Mantel Test, R 2 = 0.038, FDR‐corrected p value = 0.32). Together with the AMOVA results (Table 1) showing bigger among‐group differences for mtDNA than for the NRY, these results indicate that mtDNA tends to be geographically more localized than the NRY. To exclude the impact of the East African, Bantu, and European haplogroups, we performed the Mantel Test between pairwise geographic distances and mtDNA and NRY Φ ST distances from the AU‐NBZ dataset. The Mantel Test on this data set still did not show a significant correlation between NRY Φ ST distances and geography (Mantel Test, R 2 = 0.266, FDR‐corrected p value = 0.12), but it did show an increase in R 2 value. In contrast, mtDNA Φ ST distances and geography showed a moderate decrease in R 2 value and the correlation was not significant anymore (Mantel Test, R 2 = 0.214, FDR‐corrected p value = 0.14).
3.1.10. NRY haplogroup E1b1b and pastoralism
The NRY haplogroup E1b1b has been suggested to have an East African origin and has been associated with the spread of pastoralism from East Africa to southern Africa (Henn et al., 2008). However, this previous study included just three populations from southern Africa (!Xuun foragers, Khwe that practice various subsistence strategies, and South African Bantu agro‐pastoralists), and therefore could not test whether the E1b1b haplogroup may have been brought to southern Africa by a pre‐Bantu pastoralist migration. Assuming that there was no particularly strong drift, no changes in subsistence strategies, and that subsequent migrants (i.e., Bantu speakers or European colonists) did not contribute to the diversity of E1b1b in southern Africa or disproportionally admix with particular local populations, we would expect to see higher diversity and higher frequencies of E1b1b in modern‐day pastoralist populations. We therefore tested if the E1b1b haplogroup was in higher frequency in the pastoralist Nama than in populations with various subsistence practices and foragers, using our larger dataset of southern African Khoisan populations that practice a variety of subsistence strategies. We calculated the proportion of E1b1b in Khoisan populations separately for the pastoralist Nama, groups practicing various subsistence strategies in recorded history (ǁAni/Buga, ǁXo, Shua, Tshwa), and for traditional forager populations (Kx'a and Tuu populations, Naro, Haiǁom, Gǀui, and Gǁana; classified according to: Widlock et al. not dated, http://dobes.mpi.nl/projects/akhoe/people/; Cashdan, 1986; Barnard, 1992; Chebanne, 2002; Dieckmann, Thiem, Dirkx, & Hays, 2014). Even though our results indicate that the pastoralist Nama have the highest frequency of E1b1b, the population proportion 95% CIs overlap between different subsistence strategy groups, and there are no significant differences in frequencies between different Khoisan subsistence groups (Supporting Information Table S8; three‐sample test for equality of proportions without continuity correction p value = 0.77). To exclude possible masking of the pre‐Bantu structure, we compared frequencies of E1b1b in groups practicing different subsistence strategies after excluding Eurasian and Bantu‐related haplogroups (i.e., E1b1a, E2, G, I, K, and R1) and Khoisan populations with high proportions of non‐autochthonous ancestry (i.e., populations with predominant non‐autochthonous uniparental ancestry regardless of treatment of B2a: ǁXo, Shua, and Damara; discussed in more detail in the section “Dominant uniparental ancestry components”). After this filtering, the difference between subsistence groups is statistically significant (Supporting Information Table S8; three‐sample test for equality of proportions without continuity correction p value = 0.0058). Populations with various subsistence strategies have significantly higher frequencies of E1b1b than foragers (two‐sample test for equality of proportions with continuity correction FDR‐corrected p value = 0.023). However, the pastoralist Nama do not have significantly different frequencies than foragers or populations practicing various subsistence strategies (two‐sample test for equality of proportions with continuity correction FDR‐corrected p values = 0.32; 1, respectively). It should be noted, however, that the sample size for pastoralists in our dataset is low, the 95% confidence intervals are wide, and that there is large variation in sample size for each of the subsistence‐based groups as well as in the number of populations included in each of the groups. Thus estimates from this analysis should be taken with caution. We also tested if the E1b1b haplogroup was in higher frequency in the pooled Khoe‐Kwadi populations than in the pooled Tuu or Kx'a populations; the difference in frequency between different Khoisan linguistic groups is not significant (Supporting Information Table S8; three‐sample test for equality of proportions without continuity correction p value = 0.16). Overall, our data thus do not provide support for a link between haplogroup E1b1b and current pastoralists or Khoe‐Kwadi speakers.
3.1.11. E1b1b STRs
To further investigate the relationships of southern and eastern African E1b1b Y‐chromosomes, we genotyped our E1b1b samples for 23 STR loci and merged these with previously published data (Berniell‐Lee et al., 2009; de Filippo et al., 2011; Gomes et al., 2010; Henn et al., 2008; Tishkoff et al., 2007). A network based on the 10 STR loci in common between the studies shows that individuals from East Africa have the highest diversity of haplotypes in the data set, while among the southern African Khoisan populations, Khoe and Kx'a speakers harbor the highest diversity of haplotypes (Figure 3a). The two eastern African foraging populations that speak languages with click consonants, the Hadza and Sandawe, are spread across the network, and they show sharing of haplotypes with southern African populations, suggesting recent gene flow or a common origin of haplotypes in these populations. Haplotypes found in Khoe populations are shared with other Khoisan groups, East African Hadza, Sandawe and Nilotic populations, and southern African Bantu, and are generally either shared or in close proximity to Sandawe haplotypes. To see if our study supports the suggestion by Henn et al. (2008) that the DYS389I‐10 allele is most likely derived, we investigated the state of the DYS389I marker in the networks. The segregation between the two clusters in the sequence‐based network of haplogroup E1b1b indicates that the DYS389I haplotype with 10 repeats is most likely ancestral (Figure 3b), contrary to previous suggestions (Henn et al., 2008). However, these findings should be taken with precaution as there is no absolute agreement between the STR‐ and sequence‐based networks. Overall, the diversity of haplotypes seen in different Khoisan populations, with multiple star‐like expansions from haplotypes in close proximity to eastern African foragers, suggests a more complex history for haplogroup E1b1b than previously suspected.
Figure 3.
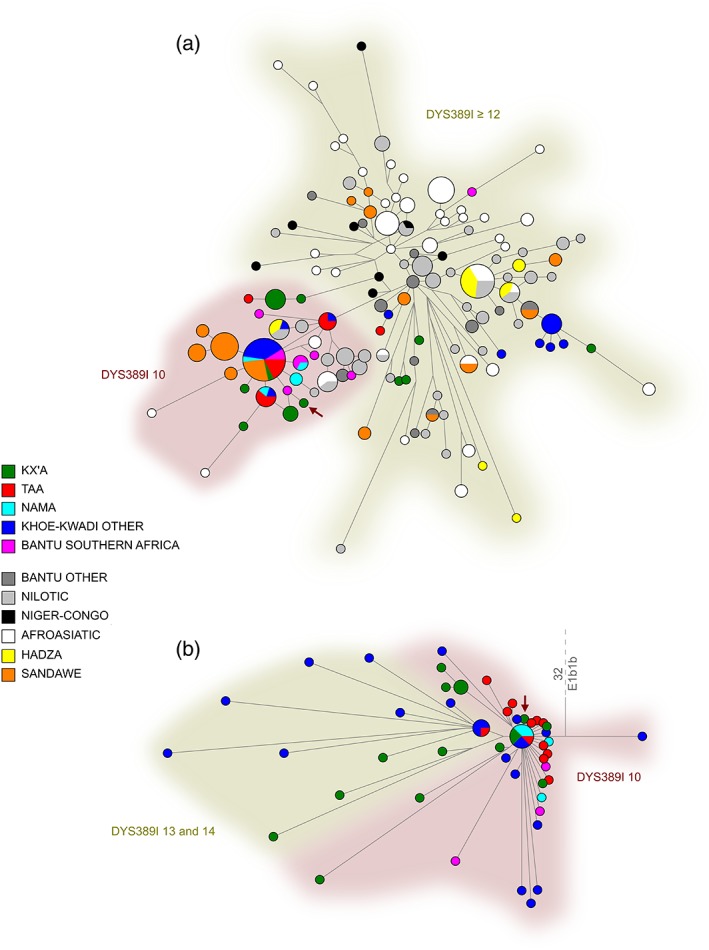
Median‐joining networks of (a) 10‐locus Y‐STR haplotypes for individuals with the NRY E1b1b haplogroup and (b) haplogroup E1b1b sequences (zoomed in from figure S12 of Barbieri et al., 2016). Shaded areas indicate the number of DYS389I repeats present in the STR haplotypes. The red arrow indicates a haplotype with 9 DYS389I repeats [Color figure can be viewed at wileyonlinelibrary.com]
3.2. Dominant uniparental ancestry components and the intensity of the sex‐biased gene flow
3.2.1. Dominant uniparental ancestry components
Most of the Khoisan populations are characterized by high frequencies of autochthonous mtDNA lineages and somewhat lower (but still relatively high) frequencies of autochthonous NRY haplogroups (Figure 4 and Supporting Information Table S6). However, a different pattern is observed for Kx'a and Tuu populations versus Khoe‐Kwadi‐speaking populations. All Kx'a and Tuu‐speaking populations are characterized by more than 80% autochthonous mtDNA lineages and variable frequencies of autochthonous NRY haplogroups (ranging from 23% to 91% regardless of whether B2a is treated as autochthonous or non‐autochthonous), while Khoe‐Kwadi populations are characterized by variability in the proportion of autochthonous mtDNA (ranging from 13% to 100%) and NRY haplogroups (B2a as AUT: 11%–83%; B2a as NAUT: 0%–81%), and thus are more widely dispersed across the plot (Figure 4). Bantu populations are characterized by an excess of autochthonous mtDNA haplogroups, and hence they show female‐biased gene flow from autochthonous populations, while in the Khoisan populations the sex‐biased gene flow is characterized by an excess of the non‐autochthonous Y‐chromosome haplogroups, showing male‐biased gene flow from non‐autochthonous populations.
Figure 4.
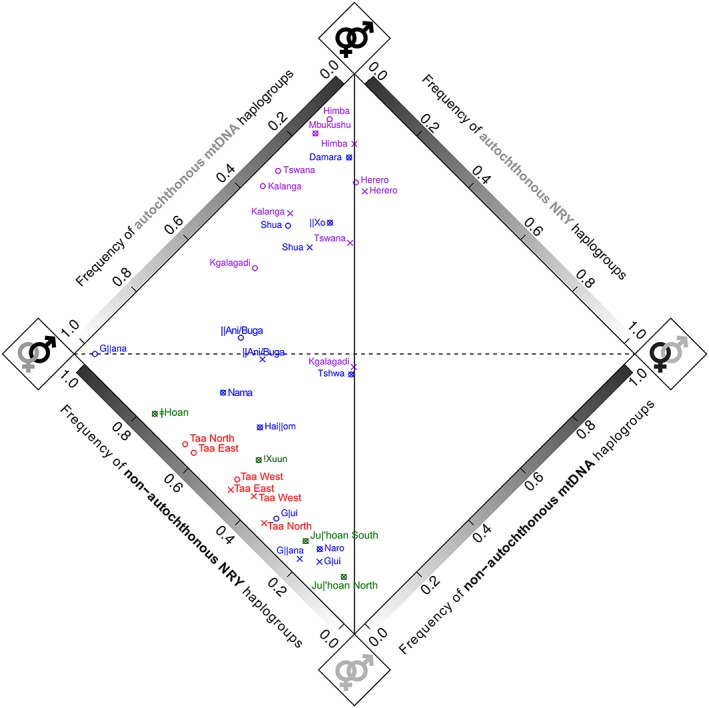
Diamond plot showing dominant uniparental ancestry components based on mtDNA and NRY haplogroup frequencies. Haplogroups of Eurasian origin are included here as non‐autochthonous haplogroups, to depict all contributions of non‐autochthonous origin to each population, but are removed from calculations of the intensity of sex‐biased gene flow (see text for further details). Language family affiliation is indicated by the color of the population name (green, Kx'a; red, Tuu; blue, Khoe‐Kwadi; purple, Bantu). The horizontal dotted black line separates populations with predominantly autochthonous haplogroups (below the line) from populations with predominantly non‐autochthonous haplogroups (above the line), based on both mtDNA and NRY haplogroup composition. The vertical black line represents equal proportions of non‐autochthonous mtDNA and NRY haplogroups (and hence no sex bias), while the distance from this line reflects the intensity of sex‐biased gene flow. The B2a haplogroup was treated separately as either autochthonous (“x”) or as non‐autochthonous (“o”) [Color figure can be viewed at wileyonlinelibrary.com]
3.2.2. Intensity of the sex‐biased gene flow
Most of the Bantu populations have moderate or no sex‐biased gene flow (with a mean of 0.137), while different Khoisan populations show varying degrees of intensity of the sex‐biased gene flow (ISBGF; with a mean of 0.386; Figure 5a,c). The Khoisan populations show significantly higher values for ISBGF than Bantu populations, regardless of the treatment of B2a as autochthonous or non‐autochthonous (Wilcoxon rank sum test with continuity correction, B2a as AUT: W = 413, FDR‐corrected p value = 0.0007, 95% CI = 0.108–0.354 (Supporting Information Figure S12c); B2a as NAUT: W = 387, FDR‐corrected p value = 0.003, 95% CI = 0.075–0.377; Figure 5c). Treating B2a as autochthonous or non‐autochthonous does not influence ISBGF (Supporting Information Figure S12) due to a strong correlation between ISBGF when B2a is treated as autochthonous versus when it is treated as non‐autochthonous (p value = 2.6 × 10−13, R 2 = 0.71; Supporting Information Figure S13). The treatment of B2a has the strongest influence on the Gǁana, who exhibit the strongest sex‐biased gene flow in the SA dataset (ISBGF = 1) when B2a is treated as a non‐autochthonous haplogroup, but have a value of 0.21 (sd of the bootstrap = 0.09) when it is treated as an autochthonous lineage. Moreover, populations in the central Kalahari and South Africa exhibit stronger ISBGF (Figure 5a), and there is a significant correlation (FDR‐corrected p value = 1.4 × 10−5, R 2 = 0.31) between latitude (north to south) and ISBGF, but not between longitude and ISBGF (Supporting Information Figure S14). The correlation between ISBGF and latitude remains significant when Khoisan and Bantu populations are analyzed separately (Bantu: B2a as NAUT: FDR‐corrected p value = 2.04 × 10−5, R 2 = 0.6; B2a as AUT: FDR‐corrected p value = 0.0034, R 2 = 0.34; Khoisan: B2a as NAUT: FDR‐corrected p value = 0.0108, R 2 = 0.33; B2a as AUT: FDR‐corrected p value = 0.0015, R 2 = 0.34; Supporting Information Figure S15). The stronger ISBGF seen in southern populations is driven mostly by higher levels of Khoisan‐specific mtDNA haplogroups in southern Bantu populations, and higher levels of Bantu‐specific NRY haplogroups in Khoisan populations (Figure 5 and Supporting Information Table S6). In Bantu populations, there is a statistically significant increase of Khoisan mtDNA lineages from north to south (FDR‐corrected p value = 7.5 × 10−7, R 2 = 0.69; Supporting Information Figure S16).
Figure 5.
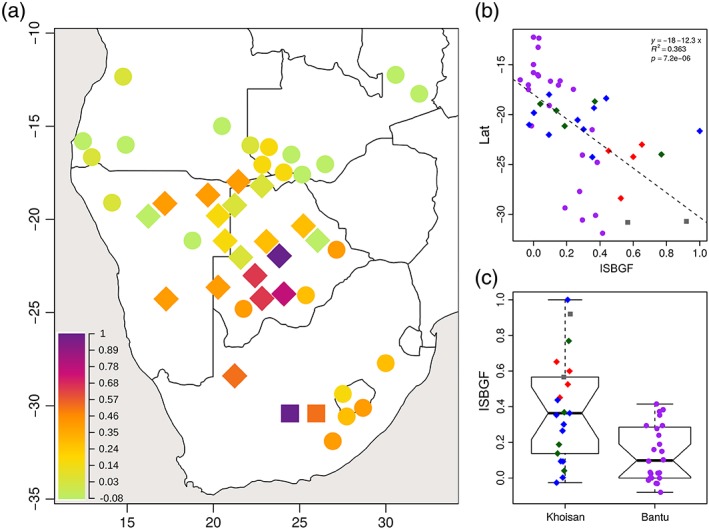
(a) Map of the intensity of sex‐biased gene flow (ISBGF) statistic, treating haplogroup B2a as a non‐autochthonous haplogroup. Dots and diamonds indicate the approximate geographic location of Bantu and Khoisan populations, respectively, and squares indicate the location of the highly admixed Karretjie and Colesberg coloured populations from South Africa. These were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (Traill, 2007; Schlebusch et al., 2011). The color indicates the ISBGF value: Values close to zero indicate equal proportions of autochthonous uniparental markers in a given population; positive values indicate a higher proportion of autochthonous mtDNA haplogroups than autochthonous Y‐chromosome haplogroups, and thus male‐biased gene flow from non‐autochthonous populations to autochthonous populations and female‐biased gene flow from autochthonous populations to non‐autochthonous populations; negative values (observed only in the Herero) indicate the opposite. (b) Correlation between latitude and ISBGF. (c) Differences in ISBGF between Khoisan and Bantu populations (Wilcoxon rank sum test with continuity correction indicates significant differences in the ISBGF means between Khoisan and Bantu populations: W = 387, FDR‐corrected p value = 0.003, 95% CI = 0.075–0.377). Different Khoisan language families are represented with different colors as in Figure 1 (with the Karretjie and Colesberg coloured populations from South Africa colored in gray) [Color figure can be viewed at wileyonlinelibrary.com]
4. DISCUSSION
4.1. Factors influencing the genetic structure of southern African populations
Southern African populations are remarkable for their genetic and cultural diversity as reflected by the wide variety of languages and different subsistence strategies. Here, we compare their maternal and paternal genetic structures and discuss the role of putative factors proposed in previous studies as important in shaping the genetic makeup of the area.
The AMOVA analysis indicates that different factors could be responsible for the current genetic structure based on the uniparental markers of southern African populations. A grouping based on ancestry components detected in autosomal data by Uren et al. (2016) is the best in explaining the variation among groups for the NRY and second‐best for mtDNA for both the entire dataset (NBZ dataset, Table 1), and for the dataset based on only autochthonous haplogroups (AU‐NBZ dataset, Supporting Information Table S7). Uren et al. (2016) based their grouping on fine‐scaled autosomal genetic structure and interpreted this as reflecting the role of geographic barriers and the ecology of the greater Kalahari Basin. To test if this interpretation is consistent with our dataset of uniparental markers and real ecotypes, we grouped populations according to terrestrial ecoregions. Our results indicate that ecology (here defined as WWF ecosystem categories in which the populations live) has very low explanatory power for both uniparental markers (WWF in Table 1). This might be an indication that the structure detected by Uren et al. does not actually reflect ecological boundaries, but rather results from a more complex mixture of geographical and historical factors. In addition, the WWF ecosystem categories are based on modern‐day data; these might not accurately reflect the prehistoric climate and vegetation patterns and thus might be poor proxies for the potential environmental factors that shaped the genetic variation seen today in southern Africa.
4.2. Fine‐scaled population structure
There are several populations that exhibit substantial frequencies of both autochthonous and non‐autochthonous uniparental markers. Many of these speak Khoe‐Kwadi languages, and they exhibit quite variable frequencies of autochthonous uniparental markers (Figure 4) and very different haplogroup compositions (Supporting Information Figure S11c,d). Some of the Khoe‐Kwadi populations are found to share identical or closely related haplotypes with Bantu populations for both mtDNA and the NRY (Supporting Information Figures S7–S9 and S17–S20), further supporting their close relationship and long history of contact with Bantu populations. We also identify several cases of mismatches between linguistic affiliation and genetic makeup, which are suggestive of language shift. We find putative examples not only of expected shifts from Khoisan to Bantu languages due to the cultural dominance of Bantu‐speaking populations, but also putative shifts from Bantu to Khoisan languages, as well as language shifts between Khoisan language families; these and other examples of discrepancies between genetic and linguistic data as well as discussions of hypotheses based on anthropological studies are discussed in more detail below.
4.2.1. Damara, Himba, and Herero
The Damara speak a Khoe language closely related to Nama, yet their current genetic makeup (Montinaro et al., 2017; Pickrell et al., 2012, 2014; Uren et al., 2016) appears to reflect shared ancestry with Bantu‐speaking populations (such as Himba and Herero), making it plausible that they have undergone language shift from a Bantu to a Khoekhoe language (Oliveira et al., 2018). These findings are supported by our data, as they appear to be genetically more similar to Bantu groups (in particular, the Himba and Herero) than to other Khoisan groups in all analyses (Figures 1, 2, and 4 and Supporting Information Figure S11). Other Khoe‐Kwadi populations reported to have high autosomal Bantu‐related ancestry (e.g., Shua, ǁXo, and to a lesser extent, Tshwa; Pickrell et al., 2014) also tend to be more similar to Bantu populations based on uniparental haplogroup composition and analyses based on Φ ST distances (Figure 2 and Supporting Information Figure S11 and Table S9). This high level of Bantu‐related ancestry, reflecting extensive admixture and/or language shift, needs to be taken into account when considering the relationships of these Khoe‐Kwadi populations.
4.2.2. Haiǁom and Nama
The Haiǁom and the Nama speak closely related Khoe languages. Barnard (1992:12) suggested that the Haiǁom originated from !Xuun speakers who shifted to the Khoekhoe language. This hypothesis is supported by the close similarity of their NRY sequences (branch 13 in Supporting Information Figure S4), and the low NRY Φ ST differentiation between the Nama, Haiǁom, and !Xuun (Figure 2 and Supporting Information Figure S11 and Table S9) as well as by the affinities between Nama and Haiǁom, and Haiǁom and !Xuun in the mtDNA (Barbieri et al., 2014). Interestingly, individual sub‐lineages of mtDNA haplogroup L3d (arrow in Supporting Information Figure S17) and NRY haplogroup E1b1a + L485 (Supporting Information Figure S7, branch 35) are found exclusively or predominantly in the Haiǁom and other Khoe‐Kwadi speakers (especially the Khwe populations from the Okavango delta: ǁAni/Buga and ǁXo), and as such they might represent remnants of lineages that were present in the proto‐Khoe‐Kwadi population that reflect ancient contact with Bantu populations.
4.2.3. ǂHoan
The ǂHoan language (nowadays referred to as ǂ'Amkoe, Gerlach, 2016; Güldemann, 2014) is geographically isolated from the other Kx'a languages with which it is related. Even though ǂHoan are linguistically related to Juǀ'hoan and !Xuun, autosomal data showed their strong relationship to neighboring Taa populations (Montinaro et al., 2017; Pickrell et al., 2012, 2014; Uren et al., 2016). The putative genetic relationship with other Kx'a speakers might be preserved in uniparental markers. However, network analysis shows that NRY sequences found in the ǂHoan are related to sequences found in the Tuu‐speaking Taa populations and in the Gǀui (Supporting Information Figures S3 and S10). The closer relationship of the ǂHoan to neighboring Khoe‐Kwadi and Tuu‐speakers than to other geographically more distant Kx'a‐speakers is additionally supported by mtDNA Φ ST distances, as they appear more similar to neighboring Khoe‐Kwadi‐speaking Gǁana and Naro, and Tuu‐speaking Taa North and Taa West, than to other Kx'a speakers (Supporting Information Table S9). The genetic data thus do not provide clear evidence for a genetic relationship with other Kx'a speakers; rather, they support a scenario of long‐term extensive contacts between the ǂHoan and surrounding groups, in good accordance with linguistic data (Gerlach, 2016).
4.2.4. Naro
Naro speak a Khoe‐Kwadi language. Their kinship system has been described as a simplified Khoe kinship system with some Juǀ'hoan features, and it has been hypothesized that they may have spoken a Kx'a language in the past and subsequently shifted to a Khoe‐Kwadi language (Barnard, 2016; Güldemann, 2008). If the hypothetical language shift in Naro was not purely a cultural process, we would expect to see genetic relationships with both Kx'a and Khoe‐Kwadi speakers. Based on both uniparental lineages, the Naro appear to be closely related to Tuu and Kx'a speakers (Supporting Information Table S9, Figure S3, branch 26 in Figure S5, dotted circle in Figure S21, dotted circle and arrow in Figure S22, haplotypes within L0d1c haplogroup indicated with arrows 2–6 in Figure S23, and haplotypes within L0d2a1 in Figure S24), which is in good accordance with previous findings based on autosomal data (Pickrell et al., 2012). However, possible genetic evidence of contact between proto‐Naro, who may have spoken a Kx'a‐related language, with Khoe‐Kwadi populations that could have contributed to the language shift in Naro, may be found in branch 11 of the network for NRY haplogroup A3b1 (Supporting Information Figure S4). This branch harbors almost exclusively haplotypes from current Khoe‐Kwadi speakers (i.e., Naro, ǁAni/Buga, and ǁXo, who all belong to the West Kalahari Khoe sub‐branch; Supporting Information Figure S1). Although most mtDNA lineages in the Naro are shared with Kx'a‐ and Tuu‐speaking groups, there are some lineages that are shared with Khoe‐Kwadi‐speaking groups, that is, one L0k lineage shared with Khwe and Gǁana (arrow in Supporting Information Figure S21), one L0d1c haplotype predominantly shared with Gǀui and East Kalahari Khoe speakers (arrow 1 in Supporting Information Figure S23), and for L0d2ab there are haplotypes shared or in close proximity to haplotypes from Haiǁom and Gǀui (Supporting Information Figure S24). Both the mtDNA and the Y‐chromosome evidence thus suggests that the putative language shift in the Naro was accompanied by some gene flow.
4.2.5. Gǁana and Kgalagadi
The Gǁana are multilingual, like most Khoisan populations; they speak both their own language (in this case, a Khoe‐Kwadi language) and a Bantu language (in this case, Kgalagadi). Barnard (1992) noted the belief present among Gǁana and the other populations that intermarriage between Khoisan females and Bantu males resulted in the founding of the Gǁana population. If this belief is indeed true, we would expect Gǁana to harbor predominantly Khoisan‐related mtDNA lineages and predominantly Bantu‐related NRY lineages. Based on their mtDNA haplogroup composition and ΦST values Gǁana are similar to other Khoisan populations, but they are a clear outlier for the NRY (Figures 1 and 4 and Supporting Information Figure S11 and Table S9). Furthermore, they are close to the Kgalagadi in the NRY‐based CA plot (Supporting Information Figure S11B). The cultural belief of extreme sex‐biased admixture is also supported by the high frequency of NRY haplogroup B2a in Gǁana (80%) and the fact that the lowest pairwise ΦST values for the NRY between Gǁana and any other population is with the Bantu‐speaking Kgalagadi (Supporting Information Table S9). Additionally, STR analyses show that there is less diversity for B2a in the Gǁana than in Bantu populations (Supporting Information Figure S6b). This mixed ancestry for the Gǁana is also reflected in the autosomal data, as they have an estimated 30%–41% Bantu‐related ancestry (Pickrell et al., 2014; Uren et al., 2016). As they have exclusively Khoisan‐related mtDNA lineages L0d and L0k (Barbieri et al., 2014; Barbieri, Vicente, et al., 2013), the probable source of autosomal Bantu ancestry is via male lineages. It is possible that all or most of the B2a haplotypes found in Gǁana came from Bantu populations, which would make the Gǁana the population that experienced the most extreme case of sex‐biased gene flow (see Figure 4 with B2a as NAUT).
The Kgalagadi show the highest proportion of autochthonous uniparental haplogroups among Bantu populations (Figure 4). Interestingly, even though some authors argue that the direct ancestors of the Tswana and Kgalagadi probably migrated to what is now Botswana as recently as 350 years ago (Kiyaga‐Mulindwa, 1993; Segobye, 1998), Barnard (1992) has suggested that the Kgalagadi are the oldest existing Bantu‐speaking inhabitants of Botswana and entered the southern part of the country probably centuries before European colonization. If the latter scenario is correct, the putative long period of cohabitation between Kgalagadi and local foraging groups in the area could explain the relatively high proportion of autochthonous uniparental haplogroups found in the Kgalagadi.
4.3. Multiple waves of sex‐biased gene flow in the context of the putative East African pastoralist migration and the Bantu expansion
Southern African populations exhibit a complex genetic makeup characterized by strong sex‐biased gene flow (Figures 4 and 5), reflecting a long history of population movements and interactions between local and incoming populations. There are at least two migration events that could have contributed to the complex and sex‐biased ancestry of Khoisan populations: the Bantu expansion and an earlier migration from eastern Africa that is putatively associated with the introduction of pastoralism (Pickrell et al., 2014).
4.3.1. Admixture with East African migrants
Archaeological and linguistic evidence has suggested a pre‐Bantu migration from eastern Africa that brought pastoralism and Khoe‐Kwadi languages (Blench, 2006; Güldemann, 2008; Mitchell, 2002; Pleurdeau et al., 2012) and thus had a significant impact on southern Africa. Although studies of different genetic markers support the demic migration model from East Africa, they vary in their conclusions concerning the impact and importance of this migration (see Introduction). For instance, the mtDNA data support very limited gene flow (Barbieri et al., 2014; Uren et al., 2016), while the lactase persistence and limited NRY data undeniably support the demic diffusion model with significant population movement (Breton et al., 2014; Henn et al., 2008; Macholdt et al., 2014, 2015; Schlebusch et al., 2012). Genome‐wide data from both ancient remains and modern populations support the demic diffusion model, but with various interpretations of its significance (Schlebusch et al., 2012, 2017; Pickrell et al., 2014; Uren et al., 2016; Montinaro et al., 2017; Skoglund et al., 2017).
One possibility is that the signal of East African ancestry in Khoisan populations was shaped by a heavily male‐mediated migration from eastern Africa (as previously proposed in Barbieri et al., 2014). Thus, it is crucial to investigate the paternal history of Khoisan populations in order to differentiate between the spread of pastoralism due to limited demic migration with more significant cultural diffusion versus a heavily male‐biased demic migration from the east that brought pastoralism. We do not find a higher frequency of the E1b1b NRY haplogroup, previously associated with the spread of pastoralism (Henn et al., 2008), in the pastoralist Nama (Supporting Information Table S8). Even though the Nama are the only sensu stricto Khoe‐speaking pastoralist population, they harbor just a subset of the E1b1b NRY diversity compared with other Khoisan populations (Figure 3 and Supporting Information Figure S10a,b). This is contrary to the expectation that diversity and frequency should be highest in the population that most probably brought pastoralism, or represents a direct descendant of such a population. If E1b1b was indeed associated with a migration of pastoralists, subsequent demographic events (e.g., migrations, drift) and/or changes in subsistence practices in southern Africa have diminished the association.
However, even though it cannot be directly associated with pastoralism, haplogroup E1b1b clearly has an eastern African origin. The close relationship of southern African E1b1b STR haplotypes to haplotypes from the two eastern African foraging populations, Hadza and Sandawe (Figure 3), indicate a common origin of haplotypes in southern and eastern Africa. This is in agreement with the linguistic hypothesis of a relationship between proto‐Khoe‐Kwadi and Sandawe (Güldemann & Elderkin, 2010). Interestingly, the diversity of haplotypes seen in different Khoisan populations, with multiple star‐like expansions from haplotypes in close proximity to eastern African foragers, suggests a more complex migration history for haplogroup E1b1b than previously suspected. This migration may have included multiple distantly related haplotypes that subsequently were sorted into different populations, and/or there may have been more than one migration event connecting eastern with southern Africa. Further studies are needed to clarify the relationships between eastern and southern African populations.
4.3.2. Bantu expansion
Our results confirm and extend previous findings concerning the enormous impact that the spread of iron‐using agro‐pastoralist populations speaking Bantu languages had on the genetic landscape of southern Africa (Barbieri et al., 2014; Beleza et al., 2005; Coelho et al., 2009; de Filippo et al., 2011; Marks et al., 2015; Oliveira et al., 2018; Pickrell et al., 2012; Skoglund et al., 2017; Wood et al., 2005). Arriving in southern Africa ∼2,000–1,200 years ago (Phillipson, 2005; Reid, Sadr, & Hanson‐James, 1998; Kinahan, 2011), the Bantu expansion resulted in a sex‐biased pattern of admixture that is characterized by a high proportion of autochthonous mtDNA lineages in agro‐pastoralist populations and a high proportion of non‐autochthonous NRY lineages in foragers (Figure 4). However, the intensity of sex‐biased admixture varies widely among populations (Figure 4) and moreover shows geographic structure (Figure 5 and Supporting Information Figure S12), indicating that this was a complex process.
Numerous differences in cultural and sex‐specific practices could have contributed to shaping the current genetic pattern seen in expanding agriculturalist Bantu‐speaking populations and local forager populations (reviewed in Heyer, Chaix, Pavard, & Austerlitz, 2012). Females from forager communities are known to be preferred by Bantu‐speaking males because of their reputation for greater fertility and the lower (if any) bride price of forager wives (Cavalli‐Sforza, 1986; Destro‐Bisol et al., 2004; Lee, 1993). The Bantu to forager flow of paternal lineages occurs if the children of such liaisons remain in the forager villages (Lee, 1993). Conversely, the flow of maternal lineages from forager to Bantu groups occurs if the children are brought up in the Bantu communities. Strong sociocultural taboos inhibit unions between forager males and Bantu females (Cavalli‐Sforza, 1986; Lee, 1993). Although this expected sex‐biased signature is stronger in Khoisan than in Bantu populations, there is considerable variation in the intensity of the sex bias among different populations (Figure 5c and Supporting Information Figure S12c), and so other factors must also play a role.
Residential practice is another cultural trait that is likely to influence the distribution of genetic variation, and thus the signal of sex‐biased gene flow. Patrilocal populations are expected to show less population differentiation for the mtDNA than the NRY because of higher rates of female migration between local groups, while the reverse is expected for matrilocal populations. Indeed, it has been observed that mtDNA is more geographically structured than the NRY in matrilocal populations (Oota, Settheetham‐Ishida, Tiwawech, Ishida, & Stoneking, 2001; Bolnick et al., 2006; Kumar et al., 2006), while patrilocal populations show contrasting patterns (Kumar et al., 2006; Langergraber et al., 2007; Oota, Settheetham‐Ishida, Tiwawech, Ishida, & Stoneking, 2001; Wilder, Kingan, Mobasher, Pilkington, & Hammer, 2004). Even though our data set consists of patrilocal Bantu populations and Khoisan populations that preferentially practice patrilocal postmarital residence after an initial period of matrilocality (and to a lesser extent neolocality; Barnard, 1992), we observe larger differences among populations for mtDNA than for the NRY (Table 1), which is the pattern characteristic for matrilocality and not for patrilocality. This deviation could be explained by a rapid male‐dominated Bantu expansion over huge geographic areas and incorporation of already geographically structured mtDNA lineages into expanding Bantu populations. Marks, Levy, Martinez‐Cadenas, Montinaro, & Capelli, (2012) showed that even though female migration is more frequent among patrilocal populations, males migrate preferentially at longer distances than females, suggesting that patrilocal residence is expected to mostly impact geographically close groups. As geographic distances between populations in our data set are mostly >200 km, which would be considered long‐range distances (Marks, et al., 2012), it is possible that the observed pattern is due to a higher migration rate of men at longer geographic distances rather than an overall higher migration rate. However, our data are insufficient for separating the effects of migration rate versus geographic distance; further studies are needed.
One of the most striking findings of our study is the increase of sex‐biased admixture from north to south. With the assumption that the initial contact between Bantu and Khoisan populations occurred in the north, the increasing ISBGF toward the south (Figure 5a,b and Supporting Information Figure S12a,b) could be interpreted as an indication that the initial contact involved less sex bias. The north to south increase in autochthonous uniparental lineages in Bantu populations (Supporting Information Figure S16) could suggest either more intense intermarriage with local populations in the south, and/or a gradual accumulation of autochthonous uniparental markers in Bantu populations toward the south as a result of interactions between the southwards migrating Bantu populations and autochthonous populations that they encountered on their way. The former seems more likely, as autochthonous lineages incorporated in Bantu populations tend to be regionally specific, for example, L0k is found mainly in the north, both in Khoisan and Bantu groups, but not in the south Bantu groups (Barbieri, Vicente, et al., 2013; Schlebusch, Lombard, & Soodyall, 2013), in contrast to what would be expected if autochthonous lineages had gradually accumulated during the southward migration of Bantu‐speaking groups. This is compatible with the Static and Moving frontiers model that implies limited assimilation of foragers into agro‐pastoralist populations during the initial expansion into a new territory, and increased likelihood of gene flow between populations with the occurrence of a static frontier once the carrying capacity of the new territory has brought the expansion of the agro‐pastoralists to a halt (Marks et al., 2015). It is also consistent with Destro‐Bisol et al.'s (2004) model that proposes the gradual establishment of social inequalities between agro‐pastoralists and foragers, resulting in asymmetric gene flow between them. During the early phase of contact between agro‐pastoralists and foragers, the survival of the newly arrived food producing societies in an unfamiliar habitat is probably heavily dependent on the knowledge of autochthonous foragers, and as such their contact might be more egalitarian. It has been suggested that Bantu languages of the Kavango‐Zambezi transfrontier region that have incorporated click consonants might have acquired them during this more egalitarian phase, when Khoisan populations had higher social status (Pakendorf, Gunnink, Sands, & Bostoen, 2017). Later, as Bantu groups became more established and less dependent on local knowledge, they became more socially dominant, and hence sex bias increased in intensity due to the establishment of sociocultural taboos (Destro‐Bisol et al., 2004). Under this interpretation, the geographic pattern would reflect an increase in sex bias due to changes in social interactions over time.
In conclusion, we have carried out a comprehensive population‐level study of matrilineal and patrilineal lineages in southern African populations, integrated within their historical and anthropological background. Discrepancies found between the linguistic and genetic relationships of Damara, Naro, ǂHoan, and Haiǁom suggest probable language shift and/or extensive contact between these and other, linguistically unrelated, populations. We find support for a migration from eastern Africa but do not find an association of NRY haplogroup E1b1b with pastoralism today, suggesting that the arrival of pastoralism was more complex than previously suspected. Our study indicates that the Bantu expansion was probably a rapid, male‐dominated expansion, during which local Khoisan females were much more likely to be absorbed into Bantu populations than Khoisan males. We detected a stronger intensity of sex‐biased gene flow in Khoisan populations than in Bantu populations via the incorporation of nonautochthonous NRY lineages into Khoisan populations. Finally, we find that the intensity of the sex‐biased gene flow increases from north to south, possibly due to the more recent arrival of Bantu populations in the south along with the gradual establishment of social inequalities between autochthonous Khoisan populations and expanding Bantu‐speaking populations. It is also possible that Bantu populations with different population histories, for example, Southern versus Southwestern Bantu speakers, may have arrived at different times. Further studies with ancient samples spanning the time frame from pre‐East African migration to post‐Bantu migration, along with further analyses of modern samples, will clarify the temporal dynamics of interactions between these migrations and the autochthonous populations.
Supporting information
Figure S1 Schematic representation of the linguistic relationships for each Khoisan language family (modified from Barbieri et al. 2013). Languages of populations included in the study are in bold. Since the time of sample collection, additional linguistic research on the Kx'a family has revealed that the language formerly referred to as ǂHoan consists of three dialects: N!aqriaxe, ǂHoan, and Sasi. The language is nowadays referred to as ǂ'Amkoe (Güldemann, 2014; Gerlach, 2016). Although the samples included under the name ǂHoan mainly stem from N!aqriaxe speakers and include only a few ǂHoan speakers, for ease of comparison with previous studies of these samples, we continue to refer to them as ǂHoan speakers.
Figure S2 Boxplots showing frequencies of major NRY haplogroups in Khoisan (light gray) and Bantu populations (dark gray). Haplogroups shaded with green are traditionally considered as Khoisan‐related, purple indicates Bantu‐related haplogroups, yellow indicates E1b1b (considered to be East African), blue are Eurasian haplogroups, and gray represents B2a, which is probably associated with both Khoisan and Bantu populations. The Bantu‐related, Eurasian, and E1b1b haplogroups are defined here as non‐autochthonous haplogroups, while Khoisan‐related haplogroups are defined as autochthonous.
Figure S3 Network of haplogroup A2 sequences (zoomed in from Supporting Information Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. The dotted circled area indicates ǂHoan haplotypes that are related to haplotypes found in the Tuu‐speaking Taa populations and in the Gǀui.
Figure S4 Network of haplogroup A3b1 sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S5 Network of haplogroup B2b sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S6 (A) Network of haplogroup B2a sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. (B) Median‐joining network of 21‐locus Y‐STR haplotypes for individuals with the NRY B2a haplogroup. The dotted circles in both networks indicate a specific sublineage found in West‐Bantu‐speaking populations (Himba, Herero, and Owambo).
Figure S7 Network of E1b1a + L485 and E1b1a1 haplogroup sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. Sequences belonging to haplogroup E1b1a8a are collapsed into the triangle for graphic purposes in this figure and shown in detail in Figure S8.
Figure S8 Network of haplogroup E1b1a8a sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S9 Network of haplogroup E2 sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al., 2016. The dashed line indicates the root of the network.
Figure S10 (A) Network of haplogroup E1b1b sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. The dotted circle indicates ǂHoan haplotypes that are related to haplotypes found in the Tuu‐speaking Taa populations and in the Gǀui. (B) Median‐joining network of 21‐locus Y‐STR haplotypes for individuals with the NRY E1b1b haplogroup. Shaded areas indicate the number of DYS389I repeats present in the STR haplotypes. The red arrow indicates a haplotype with 9 DYS389I repeats.
Figure S11 (A,B) Neighbor Joining Trees based on mtDNA and NRY ΦST distances; (C,D) Correspondence analysis (CA) plots for mtDNA and the NRY based on haplogroup frequencies, respectively.
Figure S12 (A) Map of the intensity of sex‐biased gene flow (ISBGF) statistic, treating haplogroup B2a as an autochthonous haplogroup. Dots and diamonds represent the approximate geographic location of Bantu and Khoisan populations, respectively, and squares indicate the location of the highly admixed Karretjie and Colesberg Coloured populations from South Africa. These were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (see Traill, 2007, Schlebusch et al., 2011). The color indicates the ISBGF value: values close to zero indicate equal proportions of autochthonous uniparental markers in a given population; positive values indicate a higher proportion of autochthonous mtDNA haplogroups than autochthonous Y‐chromosome haplogroups, and thus male‐biased gene flow from non‐autochthonous populations to autochthonous populations and female‐biased gene flow from autochthonous populations to non‐autochthonous populations; negative values indicate the opposite. (B) Correlation between latitude and ISBGF. (C) Differences in ISBGF between Khoisan and Bantu populations (Wilcoxon rank sum test with continuity correction indicates significant differences in the ISBGF means between Khoisan and Bantu populations: W = 413, FDR‐corrected p value = 0.0007, 95% CI = 0.108–0.354). Different Khoisan language families are represented with different colors as in Figure 1 (with the Karretjie and Colesberg Coloured populations from South Africa colored in gray).
Figure S13 Correlation between estimates of the intensity of sex‐biased gene flow (ISBGF) when haplogroup B2a is treated as non‐autochthonous (NAUT) versus autochthonous (AUT).
Figure S14 Correlation plots between ISBGF and longitude for the SA dataset when NRY haplogroup B2a is treated as non‐autochthonous (A) and as autochthonous (B).
Figure S15 Correlation plots between ISBGF and latitude and longitude for Bantu (A–D) and Khoisan (E–H) populations separately. Regardless of the treatment of B2a, a significant correlation between latitude and ISBGF was observed for both Bantu (B2a as NAUT: FDR‐corrected p value = 2.04e‐05, R 2 = 0.6; B2a as AUT: FDR‐corrected p value = 0.0034, R 2 = 0.34) and Khoisan (B2a as NAUT: FDR‐corrected p value = 0.0108, R 2 = 0.33; B2a as AUT: FDR‐corrected p value = 0.0015, R 2 = 0.34) populations.
Figure S16 Correlation plot between latitude and frequencies of Khoisan‐related mtDNA (red dots: FDR‐corrected p value = 7.5e‐07, R 2 = 0.69) and NRY (black triangles: when B2a as NAUT: FDR‐corrected p value = 0.054, R 2 = 0.15; blue squares: when B2a as AUT: FDR‐corrected p value = 0.2, R 2 = 0.03) haplogroups in Bantu populations.
Figure S17 Network of mtDNA haplogroup L3d sequences (data from Barbieri et al., 2013). The black arrow indicates a haplotype found only in Khoe‐Kwadi speakers.
Figure S18 Network of mtDNA haplogroup L3e sequences (data from Barbieri et al., 2013).
Figure S19 Network of mtDNA haplogroup L3f sequences (data from Barbieri et al., 2013).
Figure S20 Network of mtDNA haplogroup L2a sequences (data from Barbieri et al., 2013).
Figure S21 Network of mtDNA haplogroup L0k sequences (data from Barbieri et al., 2013). The dotted circle indicates Naro haplotypes that are shared with, or in close proximity to haplotypes predominantly from Kx'a speakers (Juǀ'hoan and!Xuun). The arrow indicates a Naro haplotype that is shared with haplotypes from other Khoe‐Kwadi speakers (Khwe and Gǁana).
Figure S22 Network of sequences belonging to mtDNA haplogroups L0d1 and L0d1b (data from Barbieri et al., 2013). Sequences belonging to haplogroups L0d1 and L0d1b are shown here; sequences belonging to haplogroups L0d1a and L0d1c are collapsed into triangles for graphic purposes in this figure and shown in detail in Figure S23. The black arrow indicates a Naro haplotype that is shared with predominantly Taa haplotypes. The dotted circle indicates Naro haplotypes that are shared with, or in close proximity to predominantly Juǀ'hoan haplotypes.
Figure S23 Network of sequences belonging to mtDNA haplogroups L0d1a and L0d1c (data from Barbieri et al., 2013). The dashed line indicates the root of the network. Arrow 1 indicates Naro haplotypes that are predominantly shared with haplotypes from other Khoe‐Kwadi speakers. Arrows 2–5 indicate Naro haplotypes that are shared predominantly with haplotypes from Tuu and/or Kx'a speakers. Arrow 6 indicates a Naro haplotype that is shared with predominantly Juǀ'hoan and Haiǁom haplotypes.
Figure S24 Network of mtDNA haplogroup L0d2 sequences (data from Barbieri et al., 2013).
Table S2 Sample sizes for NRY and mtDNA for the NBZ and AU‐NBZ datasets. The NRY sequences from the neighboring Khwe speaking ǁAni and Buga populations were merged into a combined ǁAni/Buga population due to their low samples sizes; the mtDNA sequences were similarly merged to be directly comparable to the NRY dataset. The NBZ dataset refers to the dataset of 23 populations (17 Khoisan and six Bantu) that overlap at the level of populations between the NRY and mtDNA datasets from Namibia, Botswana and Zambia. AU‐NBZ dataset refers to the dataset of 13 populations overlapping between mtDNA and the NRY after excluding from the NBZ data set all Bantu populations, non‐autochthonous haplogroups from Khoisan populations, and Khoisan populations with sample sizes less than eight after removal of individuals with non‐autochthonous haplogroups.
Table S7 Analysis of Molecular Variance (AMOVA) for the mtDNA and NRY based on the AU‐NBZ data set. Names of different groupings are followed by the number of groups defined in brackets (see Table S5 for information on populations included in each of the groups). FDR‐corrected p values significant at the 0.05 and 0.01 levels are indicated with (*) and (**), respectively.
Table S8 Frequency of the NRY E1b1b haplogroup in Khoisan groups pooled by subsistence strategy and linguistic family before (upper part) and after (lower part) removal of individuals with Eurasian and Bantu‐related haplogroups (i.e., E1b1a, E2, G, I, K, and R1) and populations with high proportions of non‐autochthonous ancestry (i.e., populations with predominant non‐autochthonous uniparental ancestry regardless of treatment of B2a, as discussed in more detail in the section “Dominant uniparental ancestry components”). The number of populations that were used for the calculations, and the number of individuals that were used to calculate frequencies, are denoted as N and n, respectively. Populations were assigned to subsistence strategies according to: Widlock et al. not dated, http://dobes.mpi.nl/projects/akhoe/people/; Cashdan, 1986; Barnard, 1992; Chebanne, 2002; Dieckmann et al., 2014. Pastoralist: Nama; various subsistence: ǁAni/Buga, ǁXo, Shua, Tshwa, and Damara; and foragers: all Kx'a and Tuu populations, Naro, Haiǁom, Gǀui, and Gǁana. Before the filtering, neither the differences in frequency between different subsistence groups (i.e., foragers, pastoralist, and various) nor the differences between linguistic families (i.e., Khoe‐Kwadi, Tuu, and Kx'a) are significant (three‐sample test for equality of proportions without continuity correction p values: 0.77 and 0.16, respectively). After the filtering the difference between linguistic families are not significant (three‐sample test for equality of proportions without continuity correction p values: 0.85), while the differences in frequencies between different subsistence groups is significant (three‐sample test for equality of proportions without continuity correction p value = 0.0058), where the category “various” subsistence strategy has a significantly higher frequency of E1b1b than foragers (two‐sample test for equality of proportions with continuity correction FDR‐corrected p value = 0.023), but pastoralists do not have a significantly different frequency than foragers (two‐sample test for equality of proportions with continuity correction FDR‐corrected p value = 0.24).
Table S1 NRY dataset with information about individual samples and sequence identifiers from Barbieri et al. (2016). *Individuals assigned to major haplogroup E1b1b are assumed to be derived for marker E‐M293 (the marker genotyped by Henn et al., 2008), even if the position was not successfully typed in all the individuals (due to low coverage in some individuals). Manual inspection confirmed that the individuals confidently assigned to the derived position for M293 are randomly distributed in all sub‐branches within the E1b1b sequence‐based network.
Table S3 Southern African Dataset (SA dataset). Dataset from southern African populations for which both mtDNA and NRY haplogroup frequencies were previously published. *The highly admixed Karretjie and Coloured Colesberg populations from South Africa were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (see Traill, 2007, Schlebusch, de Jongh, & Soodyall, 2011).
Table S4 (A) Table with genotypes for a set of 23 STRs using the PowerPlex® Y23 System for individuals assigned to E1b1b haplogroup. (B) Table with newly genotyped individuals merged with publicly available STR datasets for the E1b1b haplogroup from Africa with a total of 278 individuals with 10 overlapping STRs.
Table S5 AMOVA groupings. Detailed information about the AMOVA groupings for the NBZ (A) and AU‐NBZ (B) datasets, and populations included in each of the groups.
Table S6 Table with major NRY haplogroup frequencies. *Individuals assigned to major haplogroup E1b1b are assumed to be derived for marker E‐M293 (the marker genotyped by Henn et al., 2008), even if the position was not successfully typed in all the individuals (due to low coverage in some individuals). Manual inspection confirmed that the individuals confidently assigned to the derived position for M293 are randomly distributed in all sub‐branches within the E1b1b sequence‐based network.
Table S9 Table with mtDNA pairwise Φ ST distances (lower triangle) and NRY pairwise Φ ST distances (upper triangle).
ACKNOWLEDGMENTS
This study focuses on the genetic prehistory of populations and it does not intend to evaluate or influence the cultural identity of any group, which consists of much more than just genetic ancestry. The authors thank all sample donors for their invaluable contribution, and members of the Human Population History Group for fruitful discussions and comments. This work was carried out within the EUROCORES Programme EuroBABEL of the European Science Foundation and was supported by funds from the Deutsche Forschungsgemeinschaft and the Max Planck Society. The authors also thank the Wenner‐Gren Foundation and the LABEX ASLAN for supporting a return trip to Botswana and Namibia to share their results with the communities who participated in the study. BP is grateful to the LABEX ASLAN (ANR‐10‐LABX‐0081) of Université de Lyon for its financial support within the program “Investissements d'Avenir” (ANR‐11‐IDEX‐0007) of the French government operated by the National Research Agency (ANR).
Bajić V, Barbieri C, Hübner A, et al. Genetic structure and sex‐biased gene flow in the history of southern African populations. Am J Phys Anthropol. 2018;167:656–671. 10.1002/ajpa.23694
REFERENCES
- Bandelt, H. J. , Forster, P. , & Röhl, A. (1999). Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48. [DOI] [PubMed] [Google Scholar]
- Barbieri, C. , Butthof, A. , Bostoen, K. , & Pakendorf, B. (2013). Genetic perspectives on the origin of clicks in Bantu languages from southwestern Zambia. European Journal of Human Genetics, 21, 430–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, C. , Güldemann, T. , Naumann, C. , Gerlach, L. , Berthold, F. , Nakagawa, H. , … Pakendorf, B. (2014). Unraveling the complex maternal history of southern African Khoisan populations. American Journal of Physical Anthropology, 153, 435–448. [DOI] [PubMed] [Google Scholar]
- Barbieri, C. , Hübner, A. , Macholdt, E. , Ni, S. , Lippold, S. , Schröder, R. , … Pakendorf, B. (2016). Refining the Y‐chromosome phylogeny with southern African sequences. Human Genetics, 135, 541–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, C. , Vicente, M. , Rocha, J. , Mpoloka, S. W. , Stoneking, M. , & Pakendorf, B. (2013). Ancient substructure in early mtDNA lineages of southern Africa. American Journal of Human Genetics, 92, 285–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnard, A. (1992). Hunters and herders of southern Africa: A comparative ethnography of the Khoisan peoples. Cambridge: Cambridge University Press. [Google Scholar]
- Barnard, A. (2016). Unity versus interdisciplinarity: A future for anthropology. Current Anthropology, 57, S145–S153. [Google Scholar]
- Batini, C. , Ferri, G. , Destro‐Bisol, G. , Brisighelli, F. , Luiselli, D. , Sánchez‐Diz, P. , … Capelli, C. (2011). Signatures of the preagricultural peopling processes in sub‐Saharan Africa as revealed by the phylogeography of early Y‐chromosome lineages. Molecular Biology and Evolution, 28, 2603–2613. [DOI] [PubMed] [Google Scholar]
- Beleza, S. , Gusmão, L. , Amorim, A. , Carracedo, A. , & Salas, A. (2005). The genetic legacy of western Bantu migrations. Human Genetics, 117, 366–375. [DOI] [PubMed] [Google Scholar]
- Berniell‐Lee, G. , Calafell, F. , Bosch, E. , Heyer, E. , Sica, L. , Mouguiama‐Daouda, P. , … Comas, D. (2009). Genetic and demographic implications of the Bantu expansion: Insights from human paternal lineages. Molecular Biology and Evolution, 26, 1581–1589. [DOI] [PubMed] [Google Scholar]
- Blench, R. (2006). Archaeology, language, and the African past. Lanham: AltaMira Press. [Google Scholar]
- Bolnick, D. A. , Bolnick, D. I. , & Smith, D. G. (2006). Asymmetric male and female genetic histories among native Americans from eastern North America. Molecular Biology and Evolution, 23, 2161–2174. [DOI] [PubMed] [Google Scholar]
- Breton, G. , Schlebusch, C. M. , Lombard, M. , Sjödin, P. , Soodyall, H. , & Jakobsson, M. (2014). Lactase persistence alleles reveal partial east African ancestry of southern African Khoe pastoralists. Current Biology, 24, 852–858. [DOI] [PubMed] [Google Scholar]
- Cashdan, E. (1986). Hunter‐gatherers of the northern Kalahari In Vossen R. & Keuthmann K. (Eds.), Contemporary studies on Khoisan in honour of Oswin Köhler on the occasion of his 75th birthday (pp. 145–180). Helmut Buske Verlag: Hamburg. [Google Scholar]
- Cavalli‐Sforza, L. L. (1986). African pygmies: An evaluation of the state of research In Cavalli‐Sforza L. L. (Ed.), African pygmies (pp. 361–426). Orlando, FL: Academic Press. [Google Scholar]
- Chebanne, A. M. (2002). Shifting identities in eastern Khoe: Ethnic and language endangerment. Pula: Botswana Journal of African Studies, 16, 147–157. [Google Scholar]
- Chessel, D. , Dufour, A. B. , & Thioulouse, J. (2004). The ade4 package ‐I : One‐table methods. R News, 4, 5–10. [Google Scholar]
- Coelho, M. , Sequeira, F. , Luiselli, D. , Beleza, S. , & Rocha, J. (2009). On the edge of Bantu expansions: mtDNA, Y‐chromosome and lactase persistence genetic variation in southwestern Angola. BMC Evolutionary Biology, 9, 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darriba, D. , Taboada, G. L. , Doallo, R. , & Posada, D. (2012). jModelTest 2: More models, new heuristics and parallel computing. Nature Methods, 9, 772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Filippo, C. , Barbieri, C. , Whitten, M. , Mpoloka, S. W. , Gunnarsdóttir, E. D. , Bostoen, K. , … Pakendorf, B. (2011). Y‐chromosomal variation in sub‐Saharan Africa: Insights into the history of Niger‐Congo groups. Molecular Biology and Evolution, 28, 1255–1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Filippo, C. , Heyn, P. , Barham, L. , Stoneking, M. , & Pakendorf, B. (2010). Genetic perspectives on forager‐farmer interaction in the Luangwa valley of Zambia. American Journal of Physical Anthropology, 141, 382–394. [DOI] [PubMed] [Google Scholar]
- Deacon, H. J. , & Deacon, J. (1999). Human beginnings in South Africa: Uncovering the secrets of the stone age. Walnut Creek, CA: AltaMira Press. [Google Scholar]
- Destro‐Bisol, G. , Donati, F. , Coia, V. , Boschi, I. , Verginelli, F. , Caglià, A. , … Capelli, C. (2004). Variation of female and male lineages in sub‐Saharan populations: The importance of sociocultural factors. Molecular Biology and Evolution, 21, 1673–1682. [DOI] [PubMed] [Google Scholar]
- Dieckmann, U. , Thiem, M. , Dirkx, E. , & Hays, J. (2014). Scraping the pot: San in Namibia two decades after independence. Windhoek: Land, Environment and Development Project of the Legal Assistance Centre and Desert Research Foundation of Namibia. [Google Scholar]
- Excoffier, L. , Laval, G. , & Schneider, S. (2005). Arlequin (version 3.0): An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50. [PMC free article] [PubMed] [Google Scholar]
- Excoffier, L. , Smouse, P. E. , & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach, L. (2016). N!Aqriaxe ‐ the phonology of and endangered language of Botswana. Wiesbaden: Harrassowitz Verlag. [Google Scholar]
- Gomes, V. , Sánchez‐Diz, P. , Amorim, A. , Carracedo, Á. , & Gusmão, L. (2010). Digging deeper into east African human Y‐chromosome lineages. Human Genetics, 127, 603–613. [DOI] [PubMed] [Google Scholar]
- Grollemund, R. , Branford, S. , Bostoen, K. , Meade, A. , Venditti, C. , & Pagel, M. (2015). Bantu expansion shows that habitat alters the route and pace of human dispersals. PNAS, 112, 13296–13301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldemann, T. (2004). Reconstruction through ‘de‐construction’: The marking of person, gender, and number in the Khoe family and Kwadi. Diachronica, 21, 251–306. [Google Scholar]
- Güldemann, T. (2005). Tuu as a language family In Güldemann T. (Ed.), Studies in Tuu (southern Khoisan). University of Leipzig Papers on Africa, languages and literatures 23 (pp. 11–30). Leipzig: Institut für Afrikanistik, Universität Leipzig. [Google Scholar]
- Güldemann, T. (2008). A linguist's view: Khoe‐Kwadi speakers as the earliest food‐producers of southern Africa. South African Humanities, 20, 93–132. [Google Scholar]
- Güldemann, T. , & Elderkin, E. D. (2010). On external genealogical relationships of the Khoe family. In: Khoisan languages and linguistics: Proceedings of the 1st international symposium January 4–8 (pp. 15–52). Köln: Rüdiger Köppe. [Google Scholar]
- Güldemann, T. (2014). “Khoisan” linguistic classification today In Güldemann T. & Fehn A. (Eds.), Beyond “Khoisan”. Historical relations in the Kalahari Basin (pp. 1–40). Amsterdam: John Benjamins Publishing Company. [Google Scholar]
- Heine, B. , & Honken, H. (2010). The Kx'a family: A new Khoisan genealogy. Journal of Asian and African Studies, 79, 5–36. [Google Scholar]
- Heinila M. 2012. Available from: http://dna.cfsna.net/HAP/Mutation_Rates.htm (accessed Feb 2016)
- Henn, B. M. , Gignoux, C. , Lin, A. A. , Oefner, P. J. , Shen, P. , Scozzari, R. , … Underhill, P. A. (2008). Y‐chromosomal evidence of a pastoralist migration through Tanzania to southern Africa. PNAS, 105, 10693–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn, B. M. , Gignoux, C. R. , Jobin, M. , Granka, J. M. , Macpherson, J. M. , Kidd, J. M. , … Feldman, M. W. (2011). Hunter‐gatherer genomic diversity suggests a southern African origin for modern humans. PNAS, 108, 5154–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer, E. , Chaix, R. , Pavard, S. , & Austerlitz, F. (2012). Sex‐specific demographic behaviours that shape human genomic variation. Molecular Ecology, 21, 597–612. [DOI] [PubMed] [Google Scholar]
- Kinahan, J. (2011). From the beginning: the archaeological evidence. In: M. Wallace. A History of Namibia: From the Beginning to 1990. London: Hurst and Company; p 15–43. [Google Scholar]
- Kiyaga‐Mulindwa, D. (1993). The iron age peoples of east‐Central Botswana In Shaw T., Sinclair P., Andah B., & Okpoko A. (Eds.), The archaeology of Africa: Food, metals and towns (pp. 386–390). New York: Routledge. [Google Scholar]
- Knight, A. , Underhill, P. A. , Mortensen, H. M. , Zhivotovsky, L. A. , Lin, A. A. , Henn, B. M. , … Mountain, J. L. (2003). African Y‐chromosome and mtDNA divergence provides insight into the history of click languages. Current Biology, 13, 464–473. [DOI] [PubMed] [Google Scholar]
- Kumar, V. , Langstieh, B. T. , Madhavi, K. V. , Naidu, V. M. , Singh, H. P. , Biswas, S. , … Reddy, B. M. (2006). Global patterns in human mitochondrial DNA and Y‐chromosome variation caused by spatial instability of the local cultural processes. PLoS Genetics, 2, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langergraber, K. E. , Siedel, H. , Mitani, J. C. , Wrangham, R. W. , Reynolds, V. , Hunt, K. , & Vigilant, L. (2007). The genetic signature of sex‐biased migration in Patrilocal chimpanzees and humans. PLoS One, 2, e973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, R. B. (1993). The Dobe Ju/hoansi. (2nd ed.). Fort Worth, TX, USA: Harcourt Collage Publishers. [Google Scholar]
- Macholdt, E. , Lede, V. , Barbieri, C. , Mpoloka, S. W. , Chen, H. , Slatkin, M. , … Stoneking, M. (2014). Tracing pastoralist migrations to southern Africa with lactase persistence alleles. Current Biology, 24, 875–879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macholdt, E. , Slatkin, M. , Pakendorf, B. , & Stoneking, M. (2015). New insights into the history of the C‐14010 lactase persistence variant in eastern and southern Africa. American Journal of Physical Anthropology, 156, 661–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks, S. J. , Levy, H. , Martinez‐Cadenas, C. , Montinaro, F. , & Capelli, C. (2012). Migration distance rather than migration rate explains genetic diversity in human patrilocal groups. Molecular Ecology, 21, 4958–4969. [DOI] [PubMed] [Google Scholar]
- Marks, S. J. , Montinaro, F. , Levy, H. , Brisighelli, F. , Ferri, G. , Bertoncini, S. , … Capelli, C. (2015). Static and moving frontiers: The genetic landscape of southern African Bantu‐speaking populations. Molecular Biology and Evolution, 32, 29–43. [DOI] [PubMed] [Google Scholar]
- Mitchell, P. (2002). The archaeology of southern Africa. Cambridge: Cambridge University Press. [Google Scholar]
- Montinaro, F. , Busby, G. B. J. , Gonzalez‐Santos, M. , Oosthuitzen, O. , Oosthuitzen, E. , Anagnostou, P. , … Capelli, C. (2017). Complex ancient genetic structure and cultural transitions in southern African populations. Genetics, 205, 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nenadić, O. , & Greenacre, M. (2007). Correspondence analysis in R, with two‐and three‐dimensional graphics: The ca package. Journal of Statistical Software, 20, 1–13. [Google Scholar]
- Nychka D., Furrer, R. , & Sain S. 2017. Fields: Tools for spatial data. R package version 9.6.
- Oliveira, S. , Fehn, A.‐M. , Aço, T. , Lages, F. , Gayà‐Vidal, M. , Pakendorf, B. , … Rocha, J. (2018). Matriclans shape populations: Insights from the Angolan Namib Desert into the maternal genetic history of southern Africa. American Journal of Physical Anthropology, 165, 518–535. [DOI] [PubMed] [Google Scholar]
- Olson, D. M. , Dinerstein, E. , Wikramanayake, E. D. , Burgess, N. D. , Powell, G. V. N. , Underwood, E. C. , … Kassem, K. R. (2001). Terrestrial ecoregions of the world: A new map of life on earth. Bioscience, 51, 933–938. [Google Scholar]
- Oota, H. , Settheetham‐Ishida, W. , Tiwawech, D. , Ishida, T. , & Stoneking, M. (2001). Human mtDNA and Y‐chromosome variation is correlated with matrilocal versus patrilocal residence. Nature Genetics, 29, 20–21. [DOI] [PubMed] [Google Scholar]
- Pakendorf, B. , Bostoen, K. , & de Filippo, C. (2011). Molecular perspectives on the Bantu expansion: A synthesis. Lang Dyn Chang, 1, 50–88. [Google Scholar]
- Pakendorf, B. , Gunnink, H. , Sands, B. , & Bostoen, K. (2017). Prehistoric Bantu‐Khoisan language contact. Lang Dyn Chang, 7, 1–46. [Google Scholar]
- Paradis, E. , Claude, J. , & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Phillipson, D. W. (2005). African archaeology (3rd ed.). Cambridge: Cambridge University Press. [Google Scholar]
- Pickrell, J. K. , Patterson, N. , Barbieri, C. , Berthold, F. , Gerlach, L. , Güldemann, T. , … Pakendorf, B. (2012). The genetic prehistory of southern Africa. Nature Communications, 3, 1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell, J. K. , Patterson, N. , Loh, P.‐R. , Lipson, M. , Berger, B. , Stoneking, M. , … Reich, D. (2014). Ancient west Eurasian ancestry in southern and eastern Africa. PNAS, 111, 2632–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleurdeau, D. , Imalwa, E. , Détroit, F. , Lesur, J. , Veldman, A. , Bahain, J.‐J. , & Marais, E. (2012). “Of sheep and men”: Earliest direct evidence of Caprine domestication in southern Africa at leopard cave (Erongo, Namibia). PLoS One, 7, e40340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana‐Murci, L. , Harmant, C. , Quach, H. , Balanovsky, O. , Zaporozhchenko, V. , Bormans, C. , … Behar, D. M. (2010). Strong maternal Khoisan contribution to the South African coloured population: A case of gender‐biased admixture. American Journal of Human Genetics, 86, 611–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team . 2014. R: A language and environment for statistical computing.
- Reid, D. A. M. , Sadr, K. , & Hanson‐James, N. (1998). Herding traditions In Lane P., Reid A., & Segobye A. (Eds.), Ditswa MMung: The Archaeology of Botswana (1st ed., pp. 81–100). Gaborone, Botswana: Pula Press and The Botswana Society. [Google Scholar]
- Rosa, A. , & Brehm, A. (2011). African human mtDNA phylogeography at‐a‐glance. Journal of Anthropological Sciences, 89, 25–58. [DOI] [PubMed] [Google Scholar]
- Salas, A. , Richards, M. , De la Fe, T. , Lareu, M. V. , Sobrino, B. , Sanchez‐Diz, P. , … Carracedo, A. (2002). The making of the African mtDNA landscape. American Journal of Human Genetics, 71, 1082–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch, C. M. , de Jongh, M. , & Soodyall, H. (2011). Different contributions of ancient mitochondrial and Y‐chromosomal lineages in ‘Karretjie people’ of the great Karoo in South Africa. Journal of Human Genetics, 56, 623–630. [DOI] [PubMed] [Google Scholar]
- Schlebusch, C. M. , Lombard, M. , & Soodyall, H. (2013). MtDNA control region variation affirms diversity and deep sub‐structure in populations from southern Africa. BMC Evolutionary Biology, 13, 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch, C. M. , Malmström, H. , Günther, T. , Sjödin, P. , Coutinho, A. , Edlund, H. , … Jakobsson, M. (2017). Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science, 358, 652–655. [DOI] [PubMed] [Google Scholar]
- Schlebusch, C. M. , Skoglund, P. , Sjödin, P. , Gattepaille, L. M. , Hernandez, D. , Jay, F. , … Jakobsson, M. (2012). Genomic variation in seven Khoe‐San groups reveals adaptation and complex African history. Science, 338, 374–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segobye, A. (1998). Early farming communities In Lane P., Reid A., & Segobye A. (Eds.), Ditswa Mmung: The Archaeology of Botswana (1st ed., pp. 101–114). Gaborone, Botswana: Pula Press and The Botswana Society. [Google Scholar]
- Skoglund, P. , Thompson, J. C. , Prendergast, M. E. , Mittnik, A. , Sirak, K. , Hajdinjak, M. , … Reich, D. (2017). Reconstructing prehistoric African population structure. Cell, 171, 59–71.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K. , & Nei, M. (1993). Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Molecular Biology and Evolution, 10, 512–526. [DOI] [PubMed] [Google Scholar]
- Tishkoff, S. A. , Gonder, M. K. , Henn, B. M. , Mortensen, H. , Knight, A. , Gignoux, C. , … Mountain, J. L. (2007). History of click‐speaking populations of Africa inferred from mtDNA and Y‐chromosome genetic variation. Molecular Biology and Evolution, 24, 2180–2195. [DOI] [PubMed] [Google Scholar]
- Traill, A. (1996). !Khwa‐Ka Hhouiten Hhouiten, “the rush of the storm”: The linguistic death of /Xam In Skotnes P. & South African National Gallery (Eds.), Miscast: Negotiating the presence of the Bushmen (pp. 161–183). Cape Town, South Africa: University of Cape Town Press. [Google Scholar]
- Trombetta, B. , D'Atanasio, E. , Massaia, A. , Ippoliti, M. , Coppa, A. , Candilio, F. , … Cruciani, F. (2015). Phylogeographic refinement and large scale genotyping of human Y‐chromosome haplogroup E provide new insights into the dispersal of early pastoralists in the African continent. Genome Biology and Evolution, 7, 1940–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill, P. A. , & Kivisild, T. (2007). Use of Y‐chromosome and mitochondrial DNA population structure in tracing human migrations. Annual Review of Genetics, 41, 539–564. [DOI] [PubMed] [Google Scholar]
- Underhill, P. A. , Shen, P. , Lin, A. A. , Jin, L. , Passarino, G. , Yang, W. H. , … Oefner, P. J. (2000). Y‐chromosome sequence variation and the history of human populations. Nature Genetics, 26, 358–361. [DOI] [PubMed] [Google Scholar]
- Uren, C. , Kim, M. , Martin, A. R. , Bobo, D. , Gignoux, C. R. , van Helden, P. D. , … Henn, B. M. (2016). Fine‐scale human population structure in southern Africa reflects ecogeographic boundaries. Genetics, 204, 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables, W. N. , & Ripley, B. D. (2002). Modern applied statistics with S (4th ed.). New York: Springer. [Google Scholar]
- Verdu, P. , Austerlitz, F. , Estoup, A. , Vitalis, R. , Georges, M. , Théry, S. , … Heyer, E. (2009). Origins and genetic diversity of pygmy hunter‐gatherers from western Central Africa. Current Biology, 19, 312–318. [DOI] [PubMed] [Google Scholar]
- Verdu, P. , Becker, N. S. A. , Froment, A. , Georges, M. , Grugni, V. , Quintana‐Murci, L. , … Austerlitz, F. (2013). Sociocultural behavior, sex‐biased admixture, and effective population sizes in central African pygmies and non‐pygmies. Molecular Biology and Evolution, 30, 918–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder, J. A. , Kingan, S. B. , Mobasher, Z. , Pilkington, M. M. , & Hammer, M. F. (2004). Global patterns of human mitochondrial DNA and Y‐chromosome structure are not influenced by higher migration rates of females versus males. Nature Genetics, 36, 1122–1125. [DOI] [PubMed] [Google Scholar]
- Wood, E. T. , Stover, D. A. , Ehret, C. , Destro‐Bisol, G. , Spedini, G. , McLeod, H. , … Hammer, M. F. (2005). Contrasting patterns of Y‐chromosome and mtDNA variation in Africa: Evidence for sex‐biased demographic processes. European Journal of Human Genetics, 13, 867–876. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Schematic representation of the linguistic relationships for each Khoisan language family (modified from Barbieri et al. 2013). Languages of populations included in the study are in bold. Since the time of sample collection, additional linguistic research on the Kx'a family has revealed that the language formerly referred to as ǂHoan consists of three dialects: N!aqriaxe, ǂHoan, and Sasi. The language is nowadays referred to as ǂ'Amkoe (Güldemann, 2014; Gerlach, 2016). Although the samples included under the name ǂHoan mainly stem from N!aqriaxe speakers and include only a few ǂHoan speakers, for ease of comparison with previous studies of these samples, we continue to refer to them as ǂHoan speakers.
Figure S2 Boxplots showing frequencies of major NRY haplogroups in Khoisan (light gray) and Bantu populations (dark gray). Haplogroups shaded with green are traditionally considered as Khoisan‐related, purple indicates Bantu‐related haplogroups, yellow indicates E1b1b (considered to be East African), blue are Eurasian haplogroups, and gray represents B2a, which is probably associated with both Khoisan and Bantu populations. The Bantu‐related, Eurasian, and E1b1b haplogroups are defined here as non‐autochthonous haplogroups, while Khoisan‐related haplogroups are defined as autochthonous.
Figure S3 Network of haplogroup A2 sequences (zoomed in from Supporting Information Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. The dotted circled area indicates ǂHoan haplotypes that are related to haplotypes found in the Tuu‐speaking Taa populations and in the Gǀui.
Figure S4 Network of haplogroup A3b1 sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S5 Network of haplogroup B2b sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S6 (A) Network of haplogroup B2a sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. (B) Median‐joining network of 21‐locus Y‐STR haplotypes for individuals with the NRY B2a haplogroup. The dotted circles in both networks indicate a specific sublineage found in West‐Bantu‐speaking populations (Himba, Herero, and Owambo).
Figure S7 Network of E1b1a + L485 and E1b1a1 haplogroup sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. Sequences belonging to haplogroup E1b1a8a are collapsed into the triangle for graphic purposes in this figure and shown in detail in Figure S8.
Figure S8 Network of haplogroup E1b1a8a sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network.
Figure S9 Network of haplogroup E2 sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al., 2016. The dashed line indicates the root of the network.
Figure S10 (A) Network of haplogroup E1b1b sequences (zoomed in from Figure S12 of Barbieri et al., 2016). Branches are labeled according to Figure 1 and Table S1 from Barbieri et al. (2016). The dashed line indicates the root of the network. The dotted circle indicates ǂHoan haplotypes that are related to haplotypes found in the Tuu‐speaking Taa populations and in the Gǀui. (B) Median‐joining network of 21‐locus Y‐STR haplotypes for individuals with the NRY E1b1b haplogroup. Shaded areas indicate the number of DYS389I repeats present in the STR haplotypes. The red arrow indicates a haplotype with 9 DYS389I repeats.
Figure S11 (A,B) Neighbor Joining Trees based on mtDNA and NRY ΦST distances; (C,D) Correspondence analysis (CA) plots for mtDNA and the NRY based on haplogroup frequencies, respectively.
Figure S12 (A) Map of the intensity of sex‐biased gene flow (ISBGF) statistic, treating haplogroup B2a as an autochthonous haplogroup. Dots and diamonds represent the approximate geographic location of Bantu and Khoisan populations, respectively, and squares indicate the location of the highly admixed Karretjie and Colesberg Coloured populations from South Africa. These were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (see Traill, 2007, Schlebusch et al., 2011). The color indicates the ISBGF value: values close to zero indicate equal proportions of autochthonous uniparental markers in a given population; positive values indicate a higher proportion of autochthonous mtDNA haplogroups than autochthonous Y‐chromosome haplogroups, and thus male‐biased gene flow from non‐autochthonous populations to autochthonous populations and female‐biased gene flow from autochthonous populations to non‐autochthonous populations; negative values indicate the opposite. (B) Correlation between latitude and ISBGF. (C) Differences in ISBGF between Khoisan and Bantu populations (Wilcoxon rank sum test with continuity correction indicates significant differences in the ISBGF means between Khoisan and Bantu populations: W = 413, FDR‐corrected p value = 0.0007, 95% CI = 0.108–0.354). Different Khoisan language families are represented with different colors as in Figure 1 (with the Karretjie and Colesberg Coloured populations from South Africa colored in gray).
Figure S13 Correlation between estimates of the intensity of sex‐biased gene flow (ISBGF) when haplogroup B2a is treated as non‐autochthonous (NAUT) versus autochthonous (AUT).
Figure S14 Correlation plots between ISBGF and longitude for the SA dataset when NRY haplogroup B2a is treated as non‐autochthonous (A) and as autochthonous (B).
Figure S15 Correlation plots between ISBGF and latitude and longitude for Bantu (A–D) and Khoisan (E–H) populations separately. Regardless of the treatment of B2a, a significant correlation between latitude and ISBGF was observed for both Bantu (B2a as NAUT: FDR‐corrected p value = 2.04e‐05, R 2 = 0.6; B2a as AUT: FDR‐corrected p value = 0.0034, R 2 = 0.34) and Khoisan (B2a as NAUT: FDR‐corrected p value = 0.0108, R 2 = 0.33; B2a as AUT: FDR‐corrected p value = 0.0015, R 2 = 0.34) populations.
Figure S16 Correlation plot between latitude and frequencies of Khoisan‐related mtDNA (red dots: FDR‐corrected p value = 7.5e‐07, R 2 = 0.69) and NRY (black triangles: when B2a as NAUT: FDR‐corrected p value = 0.054, R 2 = 0.15; blue squares: when B2a as AUT: FDR‐corrected p value = 0.2, R 2 = 0.03) haplogroups in Bantu populations.
Figure S17 Network of mtDNA haplogroup L3d sequences (data from Barbieri et al., 2013). The black arrow indicates a haplotype found only in Khoe‐Kwadi speakers.
Figure S18 Network of mtDNA haplogroup L3e sequences (data from Barbieri et al., 2013).
Figure S19 Network of mtDNA haplogroup L3f sequences (data from Barbieri et al., 2013).
Figure S20 Network of mtDNA haplogroup L2a sequences (data from Barbieri et al., 2013).
Figure S21 Network of mtDNA haplogroup L0k sequences (data from Barbieri et al., 2013). The dotted circle indicates Naro haplotypes that are shared with, or in close proximity to haplotypes predominantly from Kx'a speakers (Juǀ'hoan and!Xuun). The arrow indicates a Naro haplotype that is shared with haplotypes from other Khoe‐Kwadi speakers (Khwe and Gǁana).
Figure S22 Network of sequences belonging to mtDNA haplogroups L0d1 and L0d1b (data from Barbieri et al., 2013). Sequences belonging to haplogroups L0d1 and L0d1b are shown here; sequences belonging to haplogroups L0d1a and L0d1c are collapsed into triangles for graphic purposes in this figure and shown in detail in Figure S23. The black arrow indicates a Naro haplotype that is shared with predominantly Taa haplotypes. The dotted circle indicates Naro haplotypes that are shared with, or in close proximity to predominantly Juǀ'hoan haplotypes.
Figure S23 Network of sequences belonging to mtDNA haplogroups L0d1a and L0d1c (data from Barbieri et al., 2013). The dashed line indicates the root of the network. Arrow 1 indicates Naro haplotypes that are predominantly shared with haplotypes from other Khoe‐Kwadi speakers. Arrows 2–5 indicate Naro haplotypes that are shared predominantly with haplotypes from Tuu and/or Kx'a speakers. Arrow 6 indicates a Naro haplotype that is shared with predominantly Juǀ'hoan and Haiǁom haplotypes.
Figure S24 Network of mtDNA haplogroup L0d2 sequences (data from Barbieri et al., 2013).
Table S2 Sample sizes for NRY and mtDNA for the NBZ and AU‐NBZ datasets. The NRY sequences from the neighboring Khwe speaking ǁAni and Buga populations were merged into a combined ǁAni/Buga population due to their low samples sizes; the mtDNA sequences were similarly merged to be directly comparable to the NRY dataset. The NBZ dataset refers to the dataset of 23 populations (17 Khoisan and six Bantu) that overlap at the level of populations between the NRY and mtDNA datasets from Namibia, Botswana and Zambia. AU‐NBZ dataset refers to the dataset of 13 populations overlapping between mtDNA and the NRY after excluding from the NBZ data set all Bantu populations, non‐autochthonous haplogroups from Khoisan populations, and Khoisan populations with sample sizes less than eight after removal of individuals with non‐autochthonous haplogroups.
Table S7 Analysis of Molecular Variance (AMOVA) for the mtDNA and NRY based on the AU‐NBZ data set. Names of different groupings are followed by the number of groups defined in brackets (see Table S5 for information on populations included in each of the groups). FDR‐corrected p values significant at the 0.05 and 0.01 levels are indicated with (*) and (**), respectively.
Table S8 Frequency of the NRY E1b1b haplogroup in Khoisan groups pooled by subsistence strategy and linguistic family before (upper part) and after (lower part) removal of individuals with Eurasian and Bantu‐related haplogroups (i.e., E1b1a, E2, G, I, K, and R1) and populations with high proportions of non‐autochthonous ancestry (i.e., populations with predominant non‐autochthonous uniparental ancestry regardless of treatment of B2a, as discussed in more detail in the section “Dominant uniparental ancestry components”). The number of populations that were used for the calculations, and the number of individuals that were used to calculate frequencies, are denoted as N and n, respectively. Populations were assigned to subsistence strategies according to: Widlock et al. not dated, http://dobes.mpi.nl/projects/akhoe/people/; Cashdan, 1986; Barnard, 1992; Chebanne, 2002; Dieckmann et al., 2014. Pastoralist: Nama; various subsistence: ǁAni/Buga, ǁXo, Shua, Tshwa, and Damara; and foragers: all Kx'a and Tuu populations, Naro, Haiǁom, Gǀui, and Gǁana. Before the filtering, neither the differences in frequency between different subsistence groups (i.e., foragers, pastoralist, and various) nor the differences between linguistic families (i.e., Khoe‐Kwadi, Tuu, and Kx'a) are significant (three‐sample test for equality of proportions without continuity correction p values: 0.77 and 0.16, respectively). After the filtering the difference between linguistic families are not significant (three‐sample test for equality of proportions without continuity correction p values: 0.85), while the differences in frequencies between different subsistence groups is significant (three‐sample test for equality of proportions without continuity correction p value = 0.0058), where the category “various” subsistence strategy has a significantly higher frequency of E1b1b than foragers (two‐sample test for equality of proportions with continuity correction FDR‐corrected p value = 0.023), but pastoralists do not have a significantly different frequency than foragers (two‐sample test for equality of proportions with continuity correction FDR‐corrected p value = 0.24).
Table S1 NRY dataset with information about individual samples and sequence identifiers from Barbieri et al. (2016). *Individuals assigned to major haplogroup E1b1b are assumed to be derived for marker E‐M293 (the marker genotyped by Henn et al., 2008), even if the position was not successfully typed in all the individuals (due to low coverage in some individuals). Manual inspection confirmed that the individuals confidently assigned to the derived position for M293 are randomly distributed in all sub‐branches within the E1b1b sequence‐based network.
Table S3 Southern African Dataset (SA dataset). Dataset from southern African populations for which both mtDNA and NRY haplogroup frequencies were previously published. *The highly admixed Karretjie and Coloured Colesberg populations from South Africa were treated as Khoisan populations in this analysis as they are likely to be partly descended from Khoisan populations (see Traill, 2007, Schlebusch, de Jongh, & Soodyall, 2011).
Table S4 (A) Table with genotypes for a set of 23 STRs using the PowerPlex® Y23 System for individuals assigned to E1b1b haplogroup. (B) Table with newly genotyped individuals merged with publicly available STR datasets for the E1b1b haplogroup from Africa with a total of 278 individuals with 10 overlapping STRs.
Table S5 AMOVA groupings. Detailed information about the AMOVA groupings for the NBZ (A) and AU‐NBZ (B) datasets, and populations included in each of the groups.
Table S6 Table with major NRY haplogroup frequencies. *Individuals assigned to major haplogroup E1b1b are assumed to be derived for marker E‐M293 (the marker genotyped by Henn et al., 2008), even if the position was not successfully typed in all the individuals (due to low coverage in some individuals). Manual inspection confirmed that the individuals confidently assigned to the derived position for M293 are randomly distributed in all sub‐branches within the E1b1b sequence‐based network.
Table S9 Table with mtDNA pairwise Φ ST distances (lower triangle) and NRY pairwise Φ ST distances (upper triangle).


