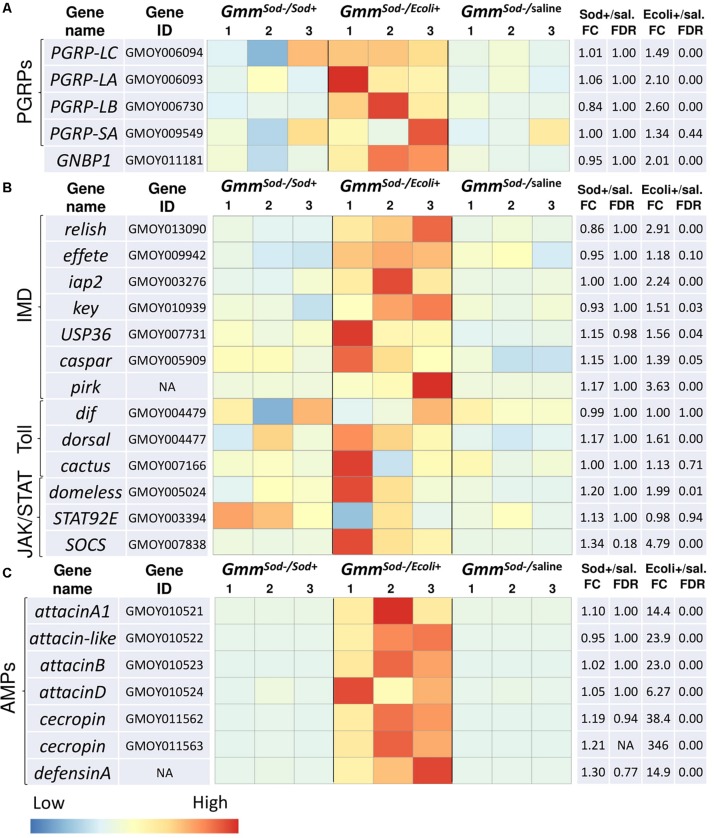
FIGURE 3.
Heat maps showing the transcriptional profiles of innate immunity-related genes obtained by RNA-seq analysis of S. glossinidius-free (GmmSod–) flies exposed to 106 CFU Sodalis glossinidius (GmmSod–/Sod+) or 105 CFU E. coli (GmmSod–/Ecoli+) and their corresponding injection-control with sterile saline (GmmSod–/saline). Bacterial exposure was performed on day 8 post-eclosion via intrathoracic microinjection as well as per os and total RNA was extracted after 48 h. Genes encoding (A) immune-related recognition proteins, (B) components of the three major immune signaling pathways; the immune deficiency (Imd)-, Toll-, and JAK/STAT-pathway, and (C) antimicrobial peptides (AMPs). Heat maps were obtained by plotting the normalized read counts scaled by row. Colors indicate the z-scores ranging from -1 (blue: low expression) to 1 (red: high expression). The biological replicates are indicated as numbers above the columns. Significantly differentially expressed genes (DEG) were defined by a p-value <0.05 and a false discovery rate (FDR; Benjamini-Hochberg adjusted p-value) <10%. Dif, dorsal-related immunity factor; Ecoli+, GmmSod–/Ecoli+; FC, fold change; GNBP1, gram-negative binding protein 1; iap2, inhibitor of apoptosis 2; key, kenny; NA, not applicable; PGRP, peptidoglycan recognition protein; sal., sterile saline-injected flies; SOCS, suppressor of cytokine signaling; Sod+, GmmSod–/Sod+; STAT92E, signal transducer and activator of transcription 92E; USP36, ubiquitin-proteasome related protein 36.