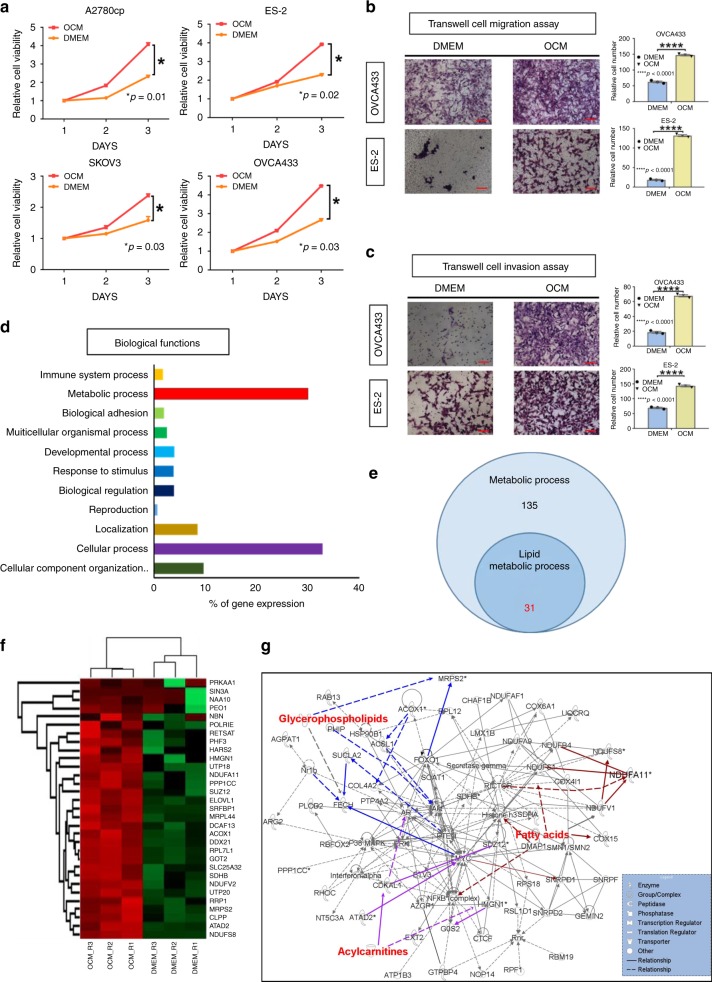
Fig. 1.
The lipid metabolic genes are frequently upregulated in ovarian cancer cells when cultured in OCM. a XTT cell proliferation assay demonstrates that treatment with OCM significantly increases cell growth in A2780cp, ES-2, SKOV3, and OVCA433 ovarian cancer cells. The relative cell viability was calculated by normalized to the mean value of day 1. b Transwell cell migration and c transwell cell invasion assays demonstrate that OCM treatment (12–24 h) promotes both cell migratory and invasive capacities in both OVCA433 and ES-2 cells. The stained cells were counted from four selected fields randomly. Representative images and quantitative results of cell migration and invasion were shown. Scale bar = 50 µm. d Ontology analysis on the altered genes detected by LC-MS/MS proteomic analysis indicates that most genes altered by OCM are associated with cellular and metabolic processes. The % of gene expression represents the % number of altered genes involved in each category of biological functions vs the total altered genes in ovarian cancer cells cultured in OCM as compared with the DMEM control. e A Venn diagram shows 31 out of 135 proteins related to metabolic processes are related to lipid metabolism. f Heatmap representation of the expression levels of the 31 proteins related to metabolism. g Ingenuity pathway analysis (IPA) depict interaction network of genes related to lipid metabolism in ovarian cancer cells. Results were presented as mean ± S.E.M. Data were analyzed by Student’s t-tests, and *p < 0.05 was considered as statistical significance