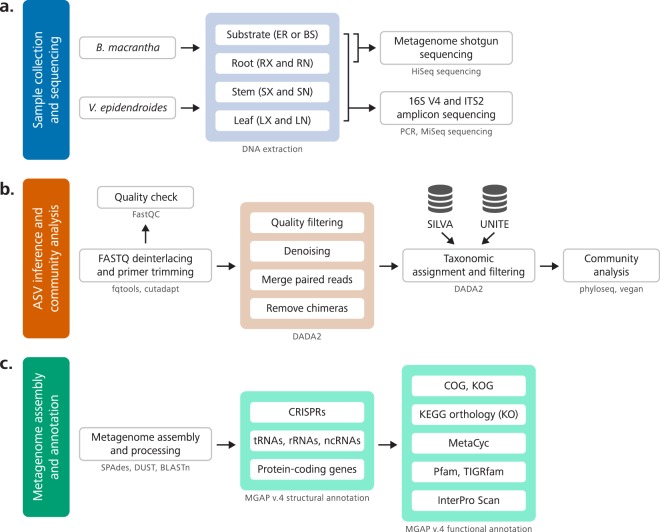
Fig. 2.
Overview of the workflows used to obtain and process the data. (a) Six individuals of both Vellozia epidendroides and Barbacenia macrantha were collected from their natural habitats and individually processed to assess the microbiomes from seven different environments through extraction of microbial DNA. The DNA extracted from three samples of four distinct communities (B. macrantha substrate, B. macrantha rhizosphere, V. epidendroides substrate and V. epidendroides rhizosphere), totaling 12 samples, was sequenced on an Illumina HiSeq platform to generate data for the metagenomic assembly. DNA from all six samples of all the assessed communities, totaling 84 samples, was used to generate 16S V4 and ITS2 amplicons, which were sequenced on an Illumina MiSeq platform. BS = bulk soil, ER = exposed rock, RX = rhizosphere, RN = endophytic root, SX = exophytic stem, SN = endophytic stem, LX = epiphytic leaf, LN = endophytic leaf. (b) The microbial community analysis started with the removal of primer sequences from the sequenced amplicons. Next, reads were denoised using the DADA2 pipeline, and the identified ASVs were assigned to bacterial and fungal taxa though comparison with the SILVA and UNITE databases, respectively. After filtering out ASVs from mitochondria and chloroplasts and low-prevalence amplicons, the phyloseq and vegan packages were used to analyze community composition. (c) The metagenomes were assembled using SPAdes software and then annotated using the standard DOE-JGI MGAP v.4 annotation pipeline. In the structural annotation step, the metagenomes were surveyed to identify CRISPRs, tRNA genes, rRNA genes, other classes of ncRNA genes and protein-coding genes. Next, the protein-coding sequences were functionally annotated and assigned to ortholog groups, metabolic pathways, chemical reactions and protein families.