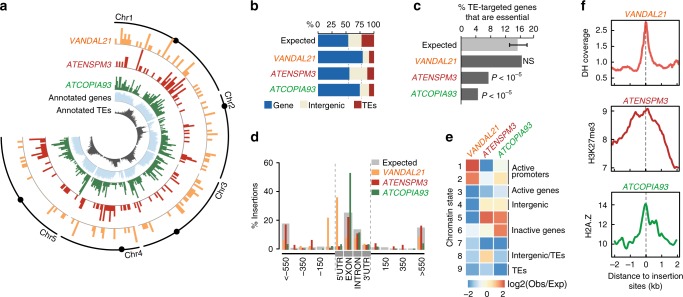
Fig. 3.
TEs exhibit strong and diverse chromatin-associated insertion biases towards genes. a Circos representation of private TE insertions detected for VANDAL21, ATENSPM3, and ATCOPIA93 within wild-type intervals. Centromere positions are indicated by black dots. b Fraction of TE insertions in genes, TEs and intergenic regions. c Fraction of essential genes among genes targeted by VANDAL21, ATENSPM3 or ATCOPIA93 in the epiRILs. Statistical significance for each comparison was obtained using the Chi-square test. d Metagene analysis showing the distribution of new insertions. e Relative abundance of insertion sites in relation to the nine chromatin states defined in A. thaliana. f Levels of DNAse hypersensitivity (DH; top panel), H3K7me3 (middle panel), and H2A.Z (bottom panel) around VANDAL21, ATENSPM3, and ATCOPIA93 insertion sites, respectively. Source data are provided as a Source Data file