Fig. 1: SCRM serves as a scaffold to recruit MAPK to interact with and modulate the stability SPCH.
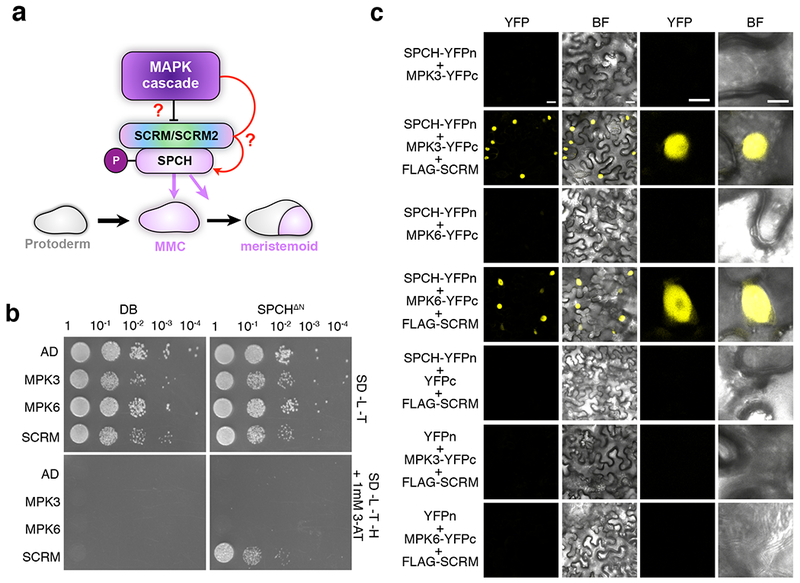
a, Schematic illustrating the influence of the MAPK cascade in regulating cell fate specification with the direct connection yet to be demonstrated (question mark). MMC, meristemoid mother cell. b, Yeast two-hybrid (Y2H) assays. Bait: the DNA binding domain alone (DB) and SPCH(ΔN-term). Prey: the activation domain alone (AD), MPK3, MPK6, SCRM and scrm-D. No detectable interaction between MPK3/MPK6 and SPCH is detected. The experiment was repeated independently three times with similar results.
c, Bimolecular Fluorescent Complementation (BiFC) assays. Shown are 3-week old N. benthamiana leaves agroinfiltrated using pairwise combinations of SPCH-YFPn and MPK3-YFPc, MPK6-YFPc along with 35S:: FLAG-SCRM (Scale bars = 25 μm). Right two panels are magnified images of a representative nucleus (scale bars = 10 μm). SPCH does not interact with MPK3/6 by itself but interacts with them only in the presence of SCRM. The experiment was repeated independently three times with similar results.
