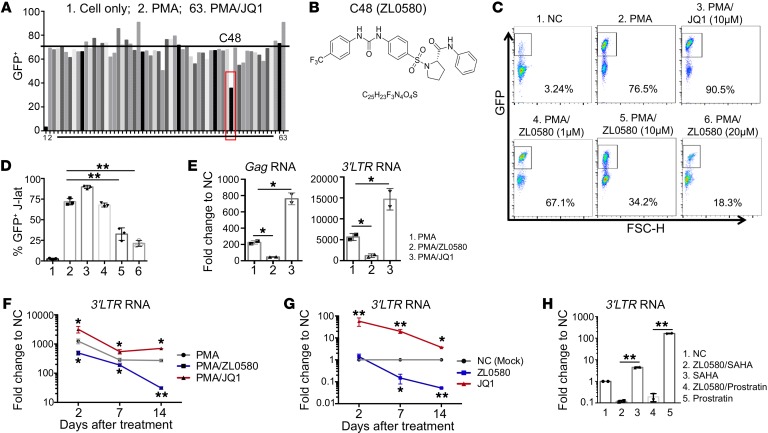
Figure 1. Discovery of a small molecule suppressing HIV in J-Lat cells.
(A) Screening of compounds (C1–C62) designed as new BRD4 modulators in J-Lat cells (10.6). Cells were stimulated with PMA (1 μg/mL) to activate HIV and treated with individual compounds (10 μM) for 24 hours (PMA/C1–C62). Cell only (NC), PMA, and PMA/JQ1 (10 μM) were included as controls (labeled as 1, 2, and 63). HIV activation was measured by flow cytometry (GFP+%). (B) Chemical structure of ZL0580. (C and D) Dose-dependent suppression of PMA-induced HIV activation by ZL0580. Cells were treated with PMA and ZL0580 (0 μM, 1 μM, 10 μM, 20 μM) for 24 hours. NC or PMA/JQ1 (10 μM) was included as a control. Representative FACS plots for GFP expression (C) and cumulative data for percentage of GFP+ in J-Lat cells of 3 experimental repeats (D) (mean ± SD) are shown. (E) Comparison of HIV transcription. HIV RNAs (Gag and 3′ LTR) were quantified by qPCR in cells 24 hours after treatment. Results are shown as fold change relative to NC. (F and G) Kinetics of ZL0580-induced HIV suppression in PMA-activated (F) or resting (G) J-Lat cells. Cells were treated as indicated for 24 hours. HIV 3′ LTR RNA was quantified on days 2, 7, and 14 after treatment. Data are shown as fold change relative to NC for each time point. Asterisks denote comparison of PMA/ZL0580 or PMA/JQ1 with PMA (F) or comparison of ZL0580 or JQ1 with NC (G). Error bars in E–G represent SD of PCR duplicate. (H) Unstimulated J-Lat cells were treated with NC or ZL0580 (10 μM), followed by stimulation with SAHA or prostratin 3 days after treatment. HIV reactivation was measured based on 3′ LTR RNA, and results are shown as fold change relative to NC. All experiments were repeated at least 3 times. *P < 0.05; **P < 0.005, 1-way ANOVA (D) and paired Student’s t test (E–H).