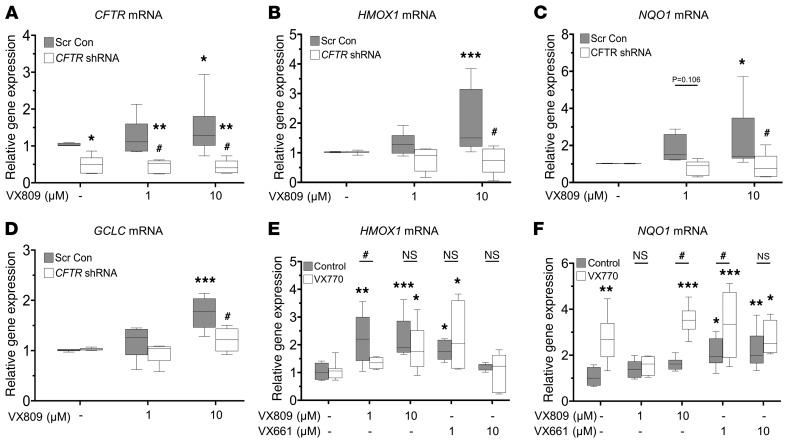
Figure 5. F508del knockdown in CFhBE cells blocks CFTR modulator–mediated activation of Nrf2.
(A–D) CFhBE cells were infected with CFTR shRNA or scrambled control (Scr Con) lentivirus for 4 days, then treated with vehicle (DMSO) control or 1–10 μM VX809 for 48 hours. Gene expression was determined for CFTR (A), HMOX1 (B), NQO1 (C), and GCLC (D), by real-time qPCR, with mRNA levels shown as fold changes versus Scr Con cells incubated with vehicle control (DMSO). Data for 4 independent experiments from 3 CF donors with 3 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 vs. Scr Con cells treated with DMSO control, or #P < 0.05 vs. Scr Con cells treated with the same dose of VX809, by mixed-effects ANOVA and Dunnett’s multiple-comparisons test. (E and F) Primary CFhBE cells were incubated with vehicle control (DMSO) or the indicated doses of VX809 or VX661, with or without 1 μM VX770, for 48 hours, and gene expression of HMOX1 (E) or NQO1 (F) was determined by qPCR as above. Data for 3–4 independent experiments from 3 CF donors with 3–4 replicates per treatment per donor are expressed as box-and-whisker plots. Horizontal bars indicate the median, box borders indicate 25th and 75th percentiles, and whiskers indicate 5th and 95th percentiles. *P < 0.05, **P < 0.01, ***P < 0.001 vs. DMSO control cells by 1-way ANOVA (Control group) or 2-way ANOVA (VX770 group) and Dunnett’s multiple-comparisons test; #P < 0.05 vs. control cells treated with the same dose of VX809/VX661 by 1-way ANOVA.