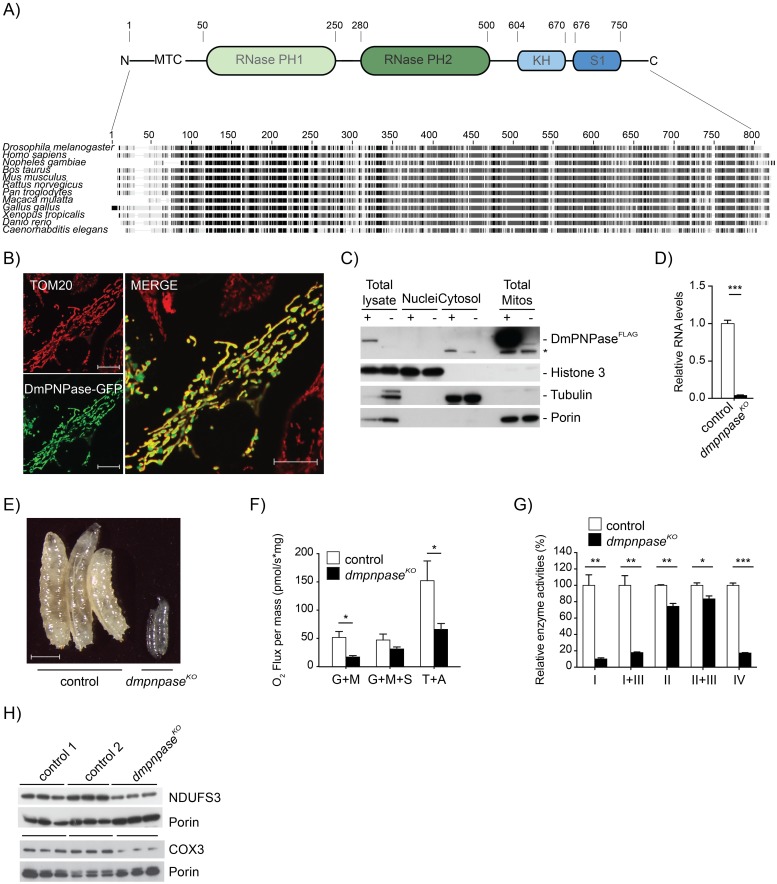
Fig 1. DmPNPase is a mitochondrial protein essential for development in Dm.
(A) ClustalW alignment of several eukaryotic PNPase sequences, as well as a schematic representation of conserved domains in DmPNPase. (B) Confocal analysis of DmPNPase-GFP fusion protein localisation in HeLa cells decorated with TOM20 (red). Scale bars: 5μm (C) Western blot analysis of nuclear, cytoplasmic and mitochondrial fractions of DmPNPase-FLAG overexpressing larvae (w;;UAS-dmpnpase-flag/daGAL4). Antibodies against the FLAG peptide, tubulin, and histone H3 were used to assess the purity of the fractions; + indicates induced expression of the FLAG tag peptide. An unspecific band is indicated by an asterisk. (D) qRT-PCR of dmpnpase transcript levels in knockout (w;;dmpnpaseKO/dmpnpaseKO) and controls (w;;) at 4 days after egg laying (AEL). Ribosomal Protein (RP) 49 transcript was used as an endogenous control. (E) Body size comparison in controls (w;;) and dmpnpaseKO larvae at 4 days AEL, scale bar size 1mm. (F) Mitochondrial oxygen consumption in dmpnpaseKO and wild type larvae (w;;). Measurements were performed on an Oroboros oxygraphy, using glutamate, malate and ADP (for complex I driven respiration), then succinate (for complex II driven respiration) and TMPD and ascorbate (for complex IV driven respiration). Error bars indicate the SEM of 9 independent experiments, each measurement was normalised to the protein content of each sample. (G) Isolated respiratory chain enzyme activities in dmpnpaseKO and control. Mitochondrial protein extracts from larvae at 4 days AEL were assessed for complex I (NADH coenzyme Q reductase), complex I+III (NADH–cytochrome c reductase), complex II (NADH cytochrome c reductase), complex III (succinate dehydrogenase), complex II+III (succinate cytochrome c reductase) and complex IV (cytochorme c oxidase). (H) Western blot analysis of mitochondrial encoded COX3 and nuclear encoded NDUFS3 respiratory chain subunits in mitochondrial protein extracts from control and dmpnpaseKO larvae at 4 days AEL. Porin was used as a loading control. (Control 1: w;;, Control 2: w;;dmpnpaseKO/TM6B). All data is represented as mean +/- SEM (***p < 0.001, **p< 0.01, *p< 0.05, n = 5).