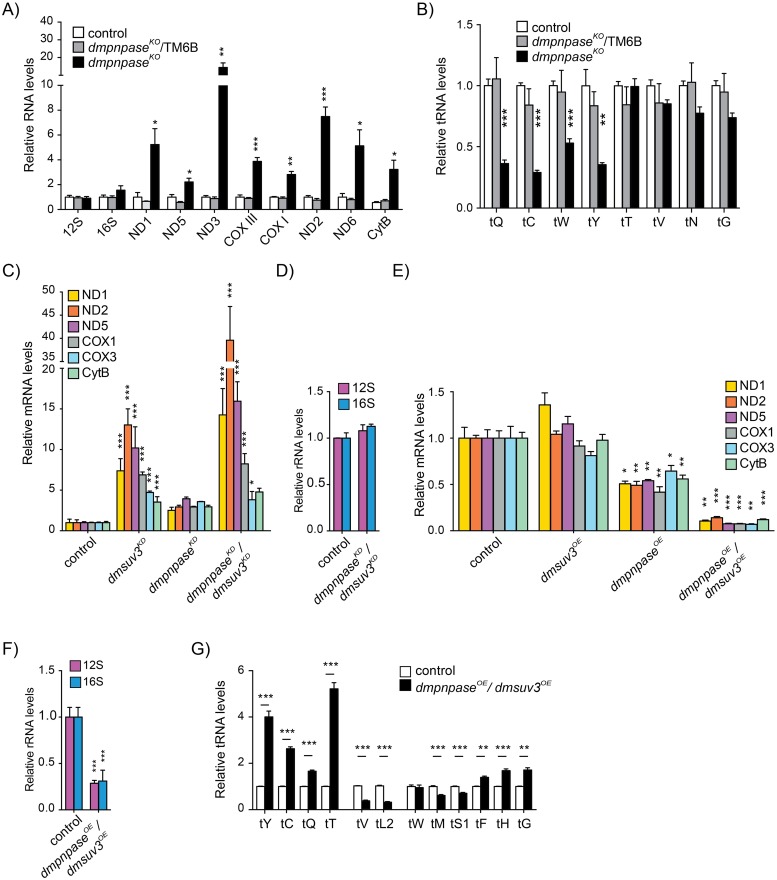
Fig 2. DmPNPase modulates the stability of mt-mRNAs.
(A) Mitochondrial mRNA steady-state levels in dmpnpaseKO and its controls (control 1: w;;daGAL4/+ and control 2: w;;dmpnpaseKO/TM6B) at 4 days AEL by qRT-PCR. RP49 transcript was used as an endogenous control. (B) Quantification of mitochondrial tRNA steady-state levels from Northern blotting analysis in S3B. Cytosolic tRNAVal was used to normalise the quantification. (C) Mitochondrial mRNA steady-state levels in DmSUV3KD (w;UAS-dmsuv3RNAi/+;daGAL4/+), dmpnpaseKO, (w;UAS-dmpnpaseRNAi;daGAL4/+) dmpnpaseKO /dmsuv3KD (w;UAS-dmsuv3RNAi/UAS-dmpnpaseRNAi;daGAL4/+) and control (w;;) larvae at 4 days AEL, as determined by qRT-PCR. RP49 transcript was used as an endogenous control. (D) Mitochondrial rRNA steady-state levels in dmpnpaseKO /dmsuv3KD determined by Northern blotting. RP49 transcript was used as an endogenous control for normalisation. (E) Mitochondrial mRNA steady-state levels in dmpnpaseOE (w;;UAS-dmpnpase/daGAL4), dmsuv3OE (w;UAS-dmsuv3/+;daGAL4/+), dmpnpaseOE/dmsuv3OE (w;UAS-dmsuv3/+;UAS-dmpnpase/daGAL4) and control (w;;) larvae at 4 days AEL, as determined by qRT-PCR. RP49 transcript was used as an endogenous control. (F) Mitochondrial rRNA steady-state levels in dmpnpaseOE/dmsuv3OE larvae determined by qRT-PCR. (G) Northern blot quantification of the steady-state levels of mitochondrial tRNAs in dmpnpaseOE/dmsuv3OE and control (w;;) larvae at 4 days AEL. Loading of the gel was normalised and quantified using a probe against cytosolic tRNAVal. All data is represented as mean +/- SEM (***p < 0.001, **p< 0.01, *p< 0.05, n = 5).