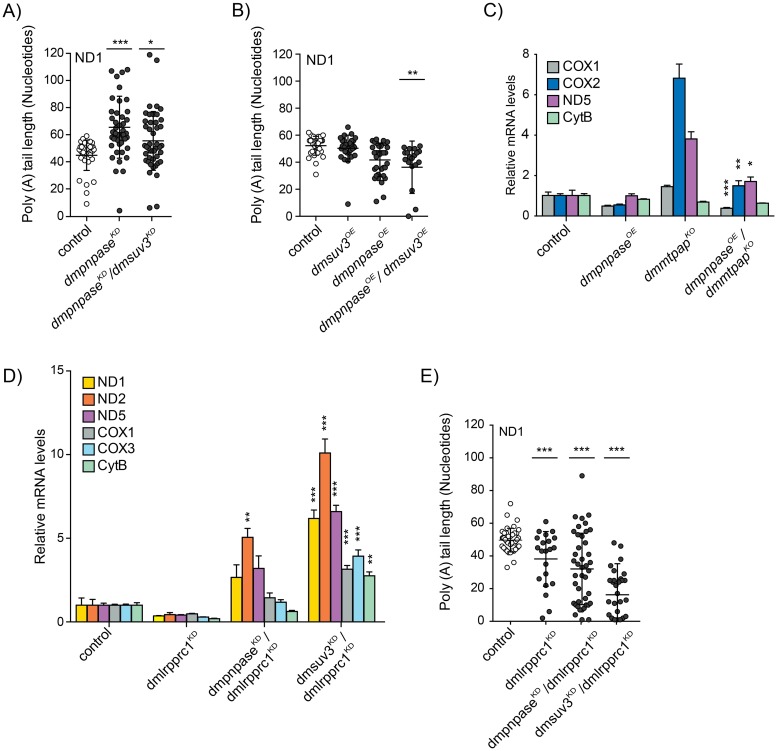
Fig 3. Polyadenylation by mtPAP is not required for degradation and sense strands are protected by LRPPRC.
(A,B) Poly(A) tail length in individually sequenced clones after 3′RACE analysis of ND1 transcripts in dmpnpaseKD, dmpnpaseKD/dmsuv3KD, dmsuv3OE, dmpnpaseOE, dmpnpaseOE/dmsuv3OE, and control (w;;) larvae at 4 days AEL. (C) Quantification of mitochondrial mRNA steady-state levels of dmpnpaseOE (dmmtpapKO/FM7;;UAS-dmpnpase/daGAL4 or FM7;;UAS-dmpnpase/daGAL4 or FM7/Y;;UAS-dmpnpase/daGAL4), dmmtpapKO (dmmtpapKO/Y;;) dmpnpaseOE/dmmtpapKO (dmmtpapKO/ Y;;UAS-dmpnpase/daGAL4), and control (w;;) larvae at 4 days AEL. Histone was used as loading control. (D) Mitochondrial mRNA steady-state levels in dmlrpprc1KD, dmlrpprc1KD/dmsuv3KD, dmlrpprc1KD/dmpnpaseKD, and control (w;;) larvae at 4 days AEL, as determined by qRT-PCR. RP49 was used as an endogenous control. (E) 3′RACE analysis of poly(A) tails of ND1 transcripts in dmlrpprc1KD (w;;UAS-bsfRNAi#1/daGAL4), dmlrpprc1KD/dmsuv3KD (w;UAS-dmsuv3RNAi/+;UAS-bsfRNAi#1/daGAL4), dmlrpprc1KD/ dmpnpaseKD (w;UAS-dmpnpaseRNAi/+;UAS-bsfRNAi#1/daGAL4), and control (w;;) larvae at 4 days AEL. All data are represented as mean +/- SEM (***p < 0.001, **p< 0.01, *p< 0.05, n = 5).