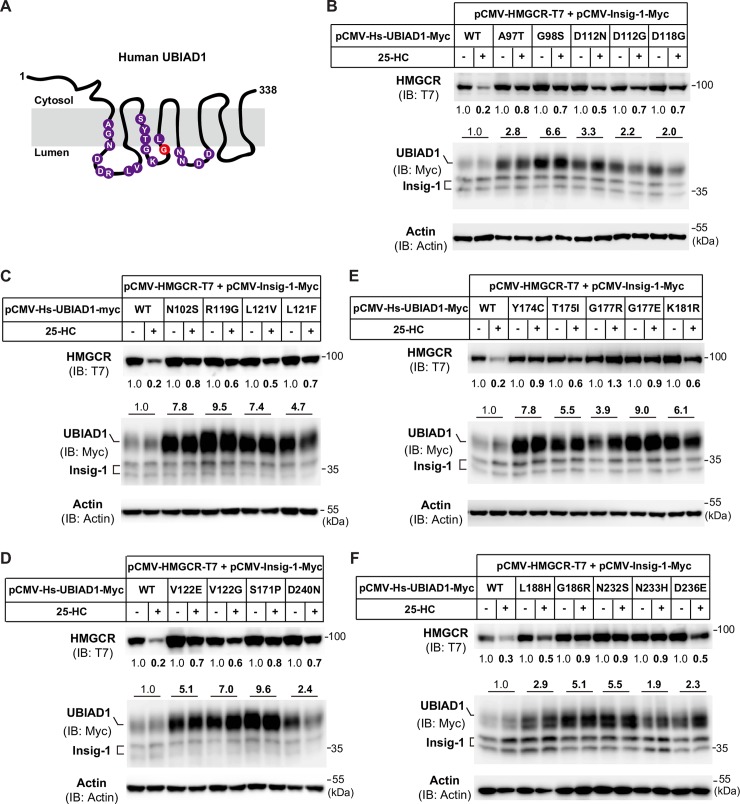
Fig 3. SCD-associated mutants of UBIAD1 block sterol-induced degradation of HMGCR.
(A) Predicted topology of human UBIAD1 protein and SCD-associated mutations of amino acids. The G186R mutation of UBIAD1 is marked with red. There are 21 different nucleotide mutations occurred in the coding sequence that altered amino acids at 19 positions. Some residues of UBIAD1 were changed to 2 different amino acids, such as D112N, D112G, G177R, G177E, V122E and V122G. (B-F) Analysis of the effects of SCD-associated mutations of UBIAD1 on the degradation of HMGCR. CHO-K1 cells were set up in 60-mm dish and transfected with 1 μg pCMV-HMGCR-T7, 0.03 μg pCMV-Insig-1-Myc and 0.1 μg indicated WT and mutant forms of pCMV-Hs-UBIAD1-Myc per dish. 6 hr after transfection, cells were washed with PBS, and depleted of sterols in medium containing 5% LPDS, 1 μM lovastatin and 50 μM mevalonate. Following depletion for 16 hr, cells were treated with or without 1 μg/ml 25-HC and 10 mM mevalonate for 6 hr. Cells were harvested for immunoblotting with indicated antibodies. Actin was a loading control. The relative protein levels of HMGCR and UBIAD1 were quantified with Image-Pro Plus 6 software, then were normalized to the Actin loading control. The normalized intensities of HMGCR without 25-HC and mevalonate treatment were defined as 1. For UBIAD1 proteins, the normalized intensities of WT UBIAD1 bands were defined as 1, and the average intensity of two UBIAD1 bands (with or without 25-HC) are shown. The experiments are repeated three times and representative data are shown.