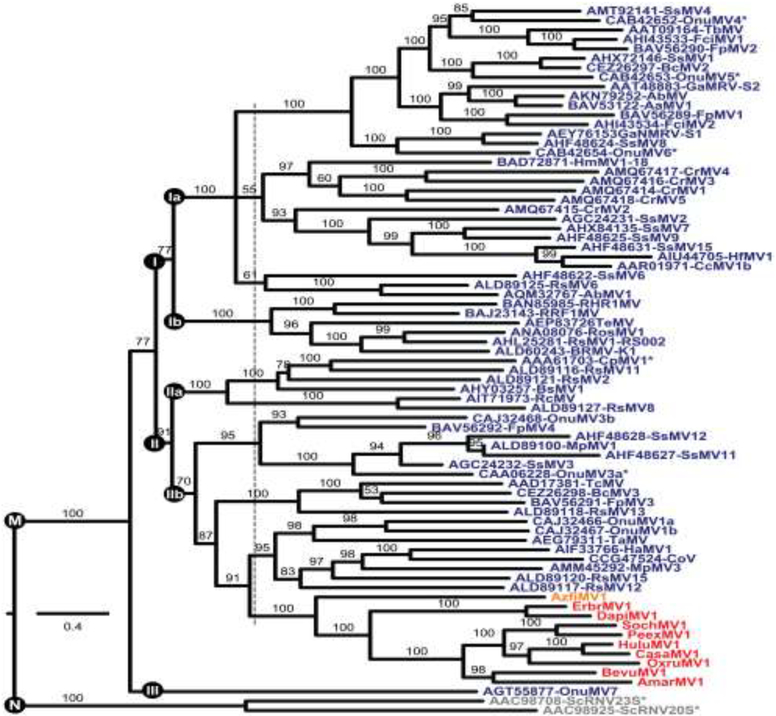
Fig. 3.
Phylogenetic tree of genus Mitovirus. Deduced RdRp sequences were aligned using MAFFT 7.310 (L-INS-i). The alignment was then analyzed using ModelFinder to determine the best-fit substitution model (VT+F+R6) and subjected to phylogenetic analysis using IQ-TREE and UFBoot as described in Materials and Methods. The site proportions and rates for the FreeRate model in this case were (0.0317,0.0275), (0.0531,0.1920), (0.1140,0.4564), (0.3158,0.8236), (0.3386,1.3151), and (0.1467,1.5775). The tree is displayed as a rectangular phylogram rooted on the branch to genus Narnavirus. Branch support values are shown in %, and branches with <50% support have been collapsed to the preceding node. Scale bar indicates average number of substitutions per alignment position. Color-coding is the same as in Fig. 2. CasaMV1, BevuMV1, DapiMV1, ErbrMV1, and PeexMV1 are represented by their reference strains. Viruses representing the 7 ratified species in genera Mitovirus (M) and Narnavirus (N) are highlighted by asterisks. Three or five apparent main clades within genus Mitovirus are labeled I–III. The dotted vertical line is explained in the main text. See Table S4 for a summary of abbreviations and GenBank numbers.