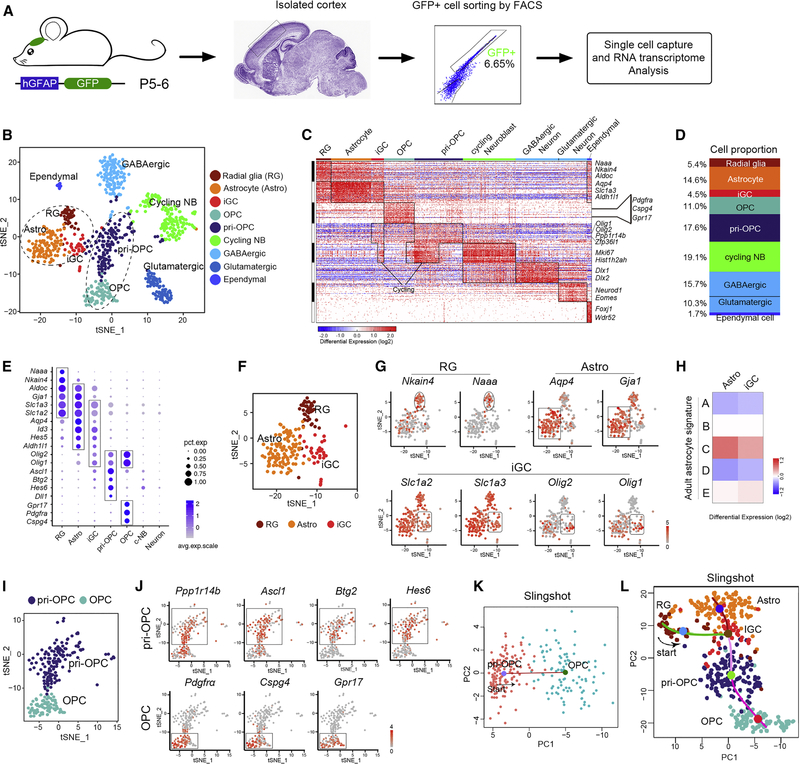
Figure 1. Unsupervised ordering of the hGFAP-GFP-derived cells reveals developmental hierarchy.
(A) Scheme for analysis of hGFAP-GFP+ cells using scRNA-seq from neonatal cortices (n=5 mice).
(B) t-SNE analysis of hGFAP-GFP+ cell clusters.
(C) Heatmap of hGFAP-GFP+ cells ordered as t-SNE (n = 815). Columns, individual cells; rows, genes.
(D) The proportions of distinct clusters among total hGFAP-GFP+ cells.
(E) Dot plot of levels of selected marker genes in subpopulations.
(F-G) t-SNE plots of F) astrocyte (Astro), radial glia (RG), and iGC and G) marker genes.
(H) Comparison of astrocyte and iGC clusters with adult astrocyte populations.
(I-J) t-SNE plot of I) OPC and pri-OPC cells and J) marker genes.
(K) Pseudo-time ordering of pri-OPCs and OPCs in hGFAP-GFP+ dataset. Red line, the predicted trajectory.
(L) Predicted lineage trajectories from RG-like cells in hGFAP-GFP+ cells.