Theoretical model suggests that gene interactions, known as epistasis, may dramatically slow down evolutionary dynamics.
Abstract
Adaptation, where a population evolves increasing fitness in a fixed environment, is typically thought of as a hill-climbing process on a fitness landscape. With a finite genome, such a process eventually leads the population to a fitness peak, at which point fitness can no longer increase through individual beneficial mutations. Instead, the ruggedness of typical landscapes due to epistasis between genes or DNA sites suggests that the accumulation of multiple mutations (via a process known as stochastic tunneling) can allow a population to continue increasing in fitness. However, it is not clear how such a phenomenon would affect long-term fitness evolution. By using a spin-glass type model for the fitness function that takes into account microscopic epistasis, we find that hopping between metastable states can mechanistically and robustly give rise to a slow, logarithmic average fitness trajectory.
INTRODUCTION
Adaptation dynamics has often been discussed in the context of fitness-parametrized landscapes, where the fitness of mutants or the fitness effects of mutations are assumed to be drawn from some distribution that could depend on the current fitness of the genotype (1–3). The resulting dynamics (e.g., fitness trajectories) depends heavily on the form of that distribution and its variation with fitness. When adopting such an approach, one often assumes an infinite genome such that a population never runs out of possible beneficial mutations.
Organisms have a finite genome, and in a large population where only beneficial mutations have a chance of fixing, one would expect the number of available beneficial mutations to deplete as cells continuously gain mutations in a constant environment (4–6). This occurs until the population eventually reaches a fitness peak where there are no possible beneficial mutations left. The fitness trajectory would therefore reach a plateau if this is the only single fitness peak in the landscape or if this process of gaining single beneficial mutations is the only way by which fitness can increase.
Interactions between loci in the genome, known as epistasis, imply that the fitness effect of a mutation depends on the states of other loci. In particular, even if individual mutations are deleterious, they could be jointly beneficial. These effects have been observed experimentally and result in the fitness landscape being rugged with many fitness peaks (7–10). In a large population, neutral and deleterious mutations cannot fix on their own, and these mutants would die out in a finite time. Escape from any local fitness peak would therefore require cells with a deleterious mutation to gain additional mutations before going extinct. This cell with multiple mutations then has a chance of fixing in the population if the mutations have a net beneficial effect. This mechanism of crossing fitness valleys has been termed stochastic tunneling and has been found to be important when the population size is sufficiently large (11, 12). At a local fitness peak, this allows the population to continue increasing in fitness. However, how this affects the form of the long-term fitness trajectory has not been studied.
In this work, we investigate how stochastic tunneling affects the average fitness trajectory of an evolving population using a spin-glass model for the fitness function and a model for the dynamics of mutation accumulation that allows a deleterious mutation to fix through fitter double mutants. We find that while the trajectory that arises within the hill-climbing regime depends on the distribution of interaction strengths between mutational sites and eventually reaches a plateau, hopping between metastable states (MSs) can robustly give rise to a slow, logarithmic fitness trajectory that is reminiscent of glassy dynamics.
RESULTS
The model
We consider a genome as a finite sequence of L sites, where each site represents a DNA base or a gene and each site can take two possible discrete states, αi = ±1 and i = 1,2, …L (Fig. 1A). The fitness, here equivalent to the exponential growth rate of a cell, depends on the states of all sites and is given by
| (1) |
where the first term sums over the independent contributions to fitness of individual sites (Fig. 1A), the second term takes into account microscopic epistasis in the form of pairwise interactions between sites [which can be considered as the lowest-order expansion in the interaction strength (13)], and Foffset is a constant whose value is chosen such that the fitness of the initial strain is 1. This is a convenient choice because we are interested in the trajectory of the relative fitness (i.e., fitness relative to the wild-type strain).
Fig. 1. The model.
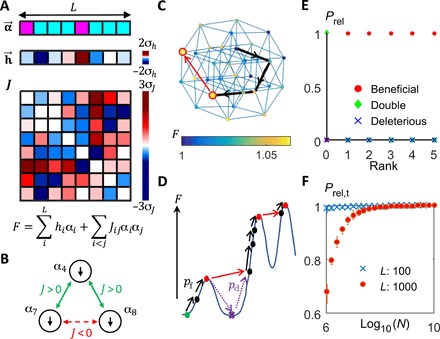
(A) The fitness of a cell is a function of the state of its genome , with each site having two possible states αi = ±1. The contributions to fitness include both the independent contributions from each of the L sites in the genome (represented by the fields ) and the pairwise interactions between sites that are captured by the symmetric matrix J. (B) The model has the feature of frustration, which can be seen by considering a triplet of sites. In this case, the 4th site is coupled positively to both the 7th and 8th sites (J4,7, J4,8 > 0) and would therefore provide a higher fitness contribution if it has the same sign as both of them. However, this would not satisfy the negative coupling between the 7th and 8th sites (J7,8 < 0). (C) Adaptation dynamics can be considered as a walk in a multidimensional genotypic space. Here, each node represents a genotypic state, with its color indicating its fitness value. Nodes corresponding to fitness peaks are enlarged and have a red border. States connected by blue lines are 1-hamming distance apart. Black arrows trace out an example of a possible adaptation trajectory to a fitness peak. The population can escape from a fitness peak via stochastic tunneling if there are states 2-hamming distance away that are higher in fitness. The red arrow shows an example of such a possible escape path. (D) The fitness landscape consists of multiple local maxima. To increase in fitness, a population away from a fitness maximum can gain a single beneficial mutation, which fixes with probability pf (black arrows). However, once the system is at a fitness maximum (red circles), it will need to acquire multiple mutations (purple dashed arrows) to transit to a state with higher fitness (red arrows). The probability of a successful double mutant emerging from a deleterious single mutant is given by pd. (E) Relative probabilities of the next successful mutation event being a single beneficial mutation (red circles), a single deleterious mutation (blue cross), or a double mutant (green diamonds). When the rank (number of available beneficial mutations) is positive, fixation of beneficial mutations dominates. At a fitness peak (rank = 0), the probability of stochastic tunneling via double mutants dominates. Each data point represents an average over 100 states on a quenched landscape (parameters: L = 200, N = 107, μ = 10−8). (F) For a given L, the relative probability of a double mutant being the next event at a fitness peak goes to 1 above some L-dependent N. Each data point represents an average over 100 randomly drawn fitness peaks, and error bars represent the interquartile range (other parameters: Nμ = 0.1, ρ = 0.05, k = 0.9, Δ = 0.05).
The strengths of the pairwise interactions are captured by the symmetric interaction matrix J (Fig. 1A) and are randomly drawn from the following distribution
| (2) |
where ρ is the average fraction of sites each site interacts with and hence determines the sparsity of the matrix. This allows the presence of frustrated interactions (Fig. 1B), which gives rise to a rugged fitness landscape (Fig. 1, C and D). We set σh = (1 − β)Δ and , where β controls the relative contribution of the field and interaction terms (for fixed ρ and L), while Δ affects the fitness effects of mutations. In our analysis, Δ is chosen such that the fitness effects of fixed mutations are, on average, of the order of a few percent (14). Because biological networks are typically sparse (15, 16), we assume that ρ is small. One could also include an additional term in Eq. 1 representing weak fully connected pairwise interactions, but this does not seem to affect the relevant structural properties of the fitness landscape as long as stronger sparse interactions are present (section SC.). For the rest of the paper, we set ρ = 0.05, but our findings hold for other values of ρ as long as it is sufficiently small (sections SC and SE).
Our model can be extended to include higher-order interaction terms if desired and, in fact, approaches that of the commonly used Kauffman’s NK model in the limit of including orders up to K (17–19), which is the number of genes each gene interacts with, and in our model is determined by ρ. However, to keep things computationally tractable even for large ρ, we will not include these higher-order interactions in our analysis. It is important to note that although we only consider pairwise interactions, each site can interact with a large number of other sites. This is in contrast to the NK model with K = 2. Similar spin-glass–type models have also been used to study other systems such as the conformational states of proteins (20, 21) and networks of neurons (22, 23). Although our model assumes that the fitness effects of independent mutations add up, mapping to the other commonly used multiplicative null model (24) gives similar trajectories (section SA).
Dynamics of mutation accumulation
We consider a population of fixed size N in a continuous culture such that growth occurs in a constant environment, and hence, the fitness landscape does not change over time. Our results also hold for batch cultures subject to the standard dilution protocol (section SF).
We model the dynamics using the Moran process, where, at each time step, a random cell is chosen to leave the population and a random cell is chosen to divide. We assume that mutations occur at a constant rate μ per cell per division and that all sites mutate with equal probability. Whenever a mutation occurs, its fitness effect is obtained using Eq. 1. To model the dynamics of how these mutations accumulate in a large population within this process, we work in the strong-selection, weak-mutation (SSWM) regime Nμ ≪ 1, where the time between successful mutations emerging is much longer than the time taken for a successful mutation to fix. This allows us to assume that if a mutation fixes, it immediately takes over the whole population. The population can then be thought of as performing an adaptive walk through a multidimensional genotypic space (Fig. 1C). Within this model, the fixation probability of a single mutant is known to be given by (25)
| (3) |
Taking ∣s∣ ≪ 1 and the large population limit, all deleterious and neutral mutations will eventually go extinct, while pf ≈ s for s ≫ 1/N.
During the time taken for deleterious/neutral mutants to go extinct, it might be possible for them to gain a second mutation such that the net fitness effect seff of the double mutant is positive. This double mutant then has a chance of fixing in the population. When the system is at a local fitness maximum, this mechanism of gaining multiple mutations is the only way the fitness can continue to increase (Fig. 1, C and D). Hence, we investigated the stability of the local peaks to multiple mutations and found that, for a small ρ, many of the fitness maxima in our model are unstable to double mutations (section SC).
We therefore take into account the probability pd that a single mutant with fitness effect s1 ≤ 0 gains a second mutation and eventually fixes, which in the large N limit and for ∣s1∣ ≪ 1 is given by (26)
| (4) |
where 〈seff〉 = 〈 max (seff,0)〉 ≪ 1, with the averaging carried out over all L possible second mutation sites. Intuitively, is the limit where the first mutant is effectively neutral and has a chance of surviving long enough (number of divisions before going extinct ) such that the probability of a successful double mutant emerging is of order 1. In the opposite limit, the first mutant is so deleterious that it would not survive for that long. Instead, nd is at most ∼1/ ∣s1∣ and pd is the probability that a successful double mutant emerges during this period (12).
This implies that if a population is large enough such that , the probability of stochastic tunneling outweighs the probability of the first deleterious mutant fixing on its own even when the first mutant is nearly neutral (first limit in Eq. 4). This condition can be satisfied together with the SSWM assumption (N ≪ 1/μ) if μ is sufficiently small μ ≪ 〈seff〉. For typical seff of a few percent, this can be easily satisfied. For the SSWM assumption to hold, a large population already implies that the mutation rate must be very small: μ ≪ 1/N. Nevertheless, we still expect stochastic tunneling to dominate the fixation of deleterious mutations at large population sizes because for any given Nμ, pf (Eq. 3) decreases with N faster than pd (Eq. 4) for s < 0.
To take into account the number of possible deleterious mutations, which is large for a long genome and can be larger than the number of possible beneficial double mutants, we constructed a typical quenched fitness landscape and, for any given state, calculated the relative probabilities of the next successful mutation event falling into one of three possible categories: (A) beneficial mutation fixing, (B) deleterious mutation fixing on its own, and (C) stochastic tunneling via a double mutant. Within our regime of interest, as long as the number of available beneficial mutations, which we refer to as the rank of the state, is positive, the next successful event must be of type A (Fig. 1E). At an MS (rank = 0), for any given L, the probability of C substantially outweighs that of B for sufficiently large N (Fig. 1F).
Because the fraction of first mutants in the population is always small, the double mutants can be assumed to be competing against the original population once it emerges. Therefore, given that a first deleterious mutant has produced a successful double mutant, the relative probabilities of the second mutation site are proportional to their fixation probabilities pf(seff).
Effective simulation model
Putting the above elements together, our effective simulation model is as follows: From any given state , we randomly draw a mutation site, calculate its fitness effect s, and accept this mutation with probability pf(s). If is a fitness peak, we allow a successful double mutant to emerge with probability pd (Eq. 4), which is obtained by considering all possible L second mutation sites and calculating the effective selection coefficients of their corresponding double mutants seff. If a successful double mutant emerges, we then draw the second mutation site randomly with weights proportional to their fixation probabilities pf(seff). For convenience, within our simulations, time is measured in terms of the number of mutational attempts (because at each time step we mutate a site and ask if the mutation successfully fixes). Although N does not explicitly enter in the simulations, we discuss in section SB the range of N for which our assumptions and results hold.
Hill-climbing regime: Relaxation toward a fitness peak
We first consider the regime where the initial state is far from a fitness peak and adaptation brings the population closer to a fitness peak.
If there is no epistasis (J = 0), there is a single well-defined peak in the fitness landscape. As mutations accumulate, the number of available beneficial mutations left, which we refer to as the rank of the system, decreases monotonically until the system eventually reaches a fitness plateau (Fig. 2A). In this case, the average fitness trajectory is uniquely defined by the distribution p(h) of h and for p(h) that follows a normal distribution, (Fig. 2B) (27).
Fig. 2. Relaxation to a fitness maximum does not generate a logarithmic fitness trajectory.

(A and B) Without epistasis (J = 0) (blue curves), the fitness reaches the global maximum quickly and follows a power law with γ = 2. With maximum epistasis (h = 0) (red curves), the fitness trajectory is slower with a power law exponent γ ≈ 0.9 but is still not as slow as a log trajectory if we do not allow escape from local fitness maxima. Here and in all other figures, time t is measured in units of number of mutational attempts (other parameters: L = 200, initial rank = 100, ρ = 0.05, Δ = 0.003).
In the presence of epistasis (J > 0), a mutation at a specific site can be beneficial or deleterious depending on the states of other sites. This is known as sign epistasis because the sign of the fitness effect of a mutation changes with the accumulation of other mutations and is responsible for a rugged fitness landscape. The rank of the system is therefore no longer guaranteed to decrease monotonically with time. By varying the relative contribution of the field and interaction terms (Eq. 1), we found that while the trajectory still follows a power law (Fig. 2, A and B), the relaxation to the first encountered local fitness maximum slows down as the relative strength of the interaction term increases (section D). Here, as in the case when J = 0, the exact form of the fitness trajectory is also sensitive to the chosen form of p(h) and p(J) but eventually reaches a fitness plateau.
Hopping between MSs
With multiple local fitness maxima, the only way for fitness to continue increasing from any of these maxima is to first gain a deleterious mutation (that does not fix by itself) followed by one or more beneficial ones (Fig. 1D). In our model, these fitness maxima are MSs that are stable to single mutations but are unstable to double mutations. Once the system escapes from an MS via a double mutant, it may enter a state with rank > 0, which is now able to gain one or more single beneficial mutations before again entering another MS, and the process repeats itself. This dynamics results in a scenario where the system spends a long time in MSs but transits between MSs relatively quickly. The phenomenon that fitness goes through periods of stasis followed by rapid jumps is known as “epochal dynamics” and has also been observed and studied in other models, such as nearly neutral holey fitness landscapes, where the epochal nature comes about from degeneracies in the genotype-to-fitness mapping (28–30).
To obtain the average fitness trajectory, instead of repeating the simulations multiple times (which would be computationally expensive because the escape events from MSs are rare and the space of possible trajectories is large), we constructed a Markov chain by mapping out all the possible states that a state can go to and by calculating the corresponding transition rates (Fig. 3A). We start the system in a randomly chosen MS (by randomly drawing a genotypic state and successively flipping sites chosen from all possible beneficial mutation sites with equal probability), assuming that the cells are already relatively close to a local fitness peak. Because double-mutation transitions are typically much slower than single-mutation transitions (Fig. 3B), we only include double-mutation transitions out of MSs, i.e., when there are no possible single-mutation transitions out of a state. The transition probability of going to a state via a single mutation at site i with effect s(i) > 0 is given by , while the probability pij of going to a double mutant state via mutations at sites i and j with net effect s(ij) is given by
| (5) |
where the first term is the probability of first gaining a mutation at i followed by j, and vice versa for the second term. The fixation probability of a mutant pf(s) and the probability of a deleterious single mutant becoming a successful double mutant pd(s1, 〈seff〉) (Eq. 4) are as defined previously, and 〈s(ik)〉 = ∑k max(s(ik),0)/L. The probability of staying in the same state after a mutation event is then 1 − ∑ipi − ∑i, jpi, j. By propagating the dynamics using this matrix (Methods), we find that the average fitness increases approximately logarithmically with time (Fig. 3C).
Fig. 3. Logarithmic fitness trajectory emerges from hopping between MSs.
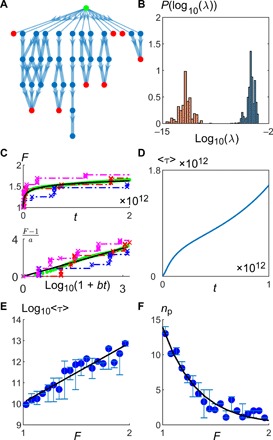
(A) The average fitness trajectory from an initial MS (green circle) was found by constructing a Markov chain that includes all possible beneficial single mutants that a non-MS (blue circles) can go to and all possible beneficial double mutants that an MS (red circles) can go to. Here, we show a subset of the whole tree, with the thickness of the arrows proportional to −1/ log(λ), where λ is the transition rate. (B) The transition rates out of an MS (red histogram) are typically much slower than those out of a non-MS (blue histogram). (C) Average fitness trajectory increases logarithmically with time. The green points are data obtained from analyzing the Markov chain, while the black line is the fit to the logarithmic function F = 1 + a log(1 + bt). The red, blue, and purple dotted lines are examples of individual trajectories, with the crosses corresponding to fixation events. (D) Average trapping time 〈τ〉 scales approximately linearly with time, showing that as the population evolves, it enters MSs that are harder and harder to get out off. (E) The average τ (blue circles) of an MS seems to increase exponentially with its fitness. The black line is a fit to a straight line. (F) The average number of escape paths from an MS np seems to decrease exponentially with its fitness. The black line is a fit to the exponential function. In both (E) and (F), the averages are taken over states in a single Markov chain, with the weight of each state proportional to the probability of encountering that state, and the error bars represent 95% confidence intervals (parameters: L = 200, μ = 10−8, β = 0.9, Δ = 0.05, ρ = 0.05).
Because the time taken to escape from an MS is much longer than the time spent between MSs, the dynamics is governed by the hopping between these MSs. This is reminiscent of Bouchaud’s trap model, where these MSs can be considered as “traps” with long trapping times τ (31), which is the average time spent in the state before transiting to another state. We also find that the average trapping time 〈τ〉 increases with time (Fig. 3D), supporting the concept that the system becomes trapped in deeper and deeper states that are harder to get out of. This is also why our model exhibits aging—the dependence on the system properties on the time from the start of the experiment—one of the hallmarks of glassy systems (section SE).
In the trap model, one assumes that the time taken to overcome energy barriers follows the Arrhenius law (31). Here, although we did not a priori assume any relationship between the trapping time τ and the fitness of a state, we find that, on average, τ increases exponentially with F (Fig. 3E). Because t ∼ τ and F ∼ log(τ), the logarithmic fitness trajectory F ∼ log(t) naturally emerges.
There are two factors that determine τ: (i) the number of escape paths np (i.e., number of possible double mutations that increases F) and (ii) the average escape rates through one of those escape paths 〈λ〉. More specifically
| (6) |
While λ depends on the fitness difference between two states and is a representative of the energy barrier between states, np governs the strength of the entropic barrier, which refers to the low probability of a favorable set of mutations occurring. We find that the exponential increase in τ with F is predominantly due to the exponential decrease in np with F (Fig. 3F) (section SE). This could be related to the exponential decrease in the number of local minima with energy in typical spin-glass energy landscapes (section SE) (32–34). Repeating the analysis with transition probabilities drawn from a uniform distribution gives a similar fitness trajectory (section SE), suggesting that it can be attributed to the following property of the fitness landscape: The number of beneficial double mutants out of an MS is negatively correlated with the fitness of the MS (section SE). This mechanism for generating a slow fitness trajectory is also robust to the choice of parameter distributions (section SE).
DISCUSSION
Biological systems in a constant environment exhibit quenched disorder, e.g., the interactions between genes are fixed and encoded in the genome, and what we typically observe experimentally are individual trajectories on a specific fitness landscape defined by a fixed set of parameters rather than the averaged trajectory over all possible realizations of the landscape. While there have been attempts to map out the fitness landscape for a few genes (35–37), repeating the process for the full landscape over all possible genotypes currently seems out of reach. In this work, we illustrate the utility of a microscopic model that explicitly takes into account specific quenched interactions between sites on a genome. We find that such a model captures realistic features of the dynamics and can provide insight into long-standing questions such as what the possible mechanisms driving long-term evolution are.
The dynamics of adaptation depends on both the fraction of available beneficial mutations in the genome and the distribution of their fitness effects. While it is common in other models to make assumptions about the beneficial mutation rate and the specific form for the fitness effect distribution, including its variation with current fitness (“macroscopic epistasis”), these aspects of the dynamics, such as the decrease in the beneficial mutation rate, emerge naturally from the microscopic model presented here.
If the number of beneficial mutations is depleted over time, as it occurs in our model, the system eventually reaches a local fitness maximum. In this case, although adaptation is commonly thought of as a hill-climbing process that only involves the accumulation of beneficial mutations, the escape from MSs via multiple mutations is crucial for the continued increase in fitness over many generations and can robustly give rise to a logarithmic fitness trajectory.
The slow fitness trajectory that emerges from hopping between MSs arises due to the ruggedness of the fitness landscape. This is reminiscent of glassy dynamics, where the existence of multiple local energy minima has also been the cause of slow relaxations in many other models of glassy systems such as spin glasses, structural glasses, electron glasses, polymers, and granular materials (38–43). However, the exact dynamics for the fitness increase in our model is different from that in these other scenarios and thus provides an alternative mechanism for how slow relaxations could arise in a real system.
A logarithmic fitness trajectory has also been known to emerge on an uncorrelated, House-of-Cards fitness landscape, where mutant fitness values are assumed to be drawn from the same distribution regardless of the fitness of the current state (1, 44). If this distribution has an exponentially decaying tail, the number of available beneficial mutations will also decrease exponentially with fitness. This has a similar flavor to the dynamics observed in our model in that the slowness of the fitness trajectory is governed by entropic factors. However, it has been shown that the number of steps to a local maximum Nsteps on an uncorrelated landscape is very short [Nsteps ∼ log(L)] (4). In contrast, the fitness landscape studied here is highly correlated (fig. S1B), and hence, both Nsteps and the number of steps between successive MSs d increase linearly with L (section SC). The presence of long-range correlations shows that the fitness values of successive MSs are still highly correlated. If each genotype was allowed to jump beyond the correlation length of the landscape by gaining many [∼O(L)] mutations in one step, the dynamics would effectively correspond to that on an uncorrelated landscape. Studies of this “long-jump” adaptation have been carried out for the evolution of NK Boolean networks, in which a logarithmic fitness trajectory has been observed (44). However, here, we found that even when considering realistic dynamics for the accumulation of single individual mutations, as long as one takes into account the phenomenon of stochastic tunneling, a slow, logarithmic average fitness trajectory can still arise on such a highly correlated landscape.
Our model can potentially be extended to account for different forms of the interaction matrix. We have so far assumed that each site interacts with a random subset of other sites, but there might be some other structure in the interaction network between genes that may affect the dynamics. For example, genes might be connected in such a way where beneficial mutations always change potential deleterious mutations into beneficial ones. This could maintain or increase the rank of the system even if it is far from an MS.
Together, our theoretical study demonstrates the utility of a microscopic model in providing a mechanistic understanding of the evolutionary dynamics and in allowing us to probe details of the system that might not be accessible in a macroscopic model. Using this approach, we explored the consequences of epistasis on fitness trajectories, both in the hill-climbing regime and after the population reaches a fitness peak, and found that hopping between MSs via stochastic tunneling can robustly give rise to a logarithmic trajectory.
METHODS
Obtaining average fitness trajectories
We obtained the probability distribution of all states in the Markov chain at time k using , where Mij is the probability of going from state j to state i in one time step (i.e., after each mutation event).
The elements of this transition matrix are determined as follows:
1) From a non-MS, the transition probability pi of going to a state via a single mutation at site i with effect s(i) > 0 is given by
2) From an MS, the probability pij of going to a double mutant state via mutations at sites i and j with net effect s(ij) is given by
where the first term is the probability of first gaining a mutation at i followed by j, and vice versa for the second term, the fixation probability of a mutant pf(s) = max(s,0), 〈s(ik)〉 = ∑k max(s(ik),0)/L, and the probability of a deleterious single mutant becoming a successful double mutant .
3) The probability of staying in the same state after a mutation event is 1 − ∑ipi − ∑i, jpi, j.
With this transition matrix, the average fitness Fk at time k can then be found from , with being the vector of fitness values of all states.
Supplementary Material
Acknowledgments
We thank B. Good, M. Desai, J. Lin, and P.-Y. Ho for useful discussions and feedback. Funding: This research was supported by the National Science Foundation through MRSEC DMR 14-20570 and PHY-1125915, the Alfred P. Sloan Foundation Research Fellowship, the Harvard Deans Competitive Fund for Promising Scholarship, the Milton Fund, the Kavli Foundation, and the Volkswagen Foundation. Author contributions: Y.G., M.V., and A.A. designed the research, performed the research, and wrote the paper. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav3842/DC1
Section SA. Choice of null model for the combined effect of multiple independent mutations
Section SB. Range of validity for population size N
Section SC. Structural features of fitness landscapes
Section SD. Relaxation to local fitness maximum
Section SE. Hopping between MSs
Section SF. Batch culture
Fig. S1. Properties of fitness landscape as a function of L.
Fig. S2. Properties of fitness peaks as a function of L when additional weak interactions
between all sites are included.
Fig. S3. Figure showing how properties of MSs vary with fitness for ρ = 0.01 (black diamonds, Foffset = −3.4), ρ = 0.025 (red circles, Foffset = −3.8), ρ = 0.05 (green triangles, Foffset = −4.2), ρ = 0.075 (blue squares, Foffset = −4.4), and ρ = 0.1 (purple crosses, Foffset = −4.5).
Fig. S4. The number of connecting MSs ns, which is the number of MSs that the system can transit to next from the current MS, correlates with the number of double mutant escape paths out of a state np.
Fig. S5. Relaxation toward a single local fitness maximum slows down with increasing degree of epistasis.
Fig. S6. Changing the distribution of fixation probabilities does not significantly change the functional form of the fitness trajectory.
Fig. S7. Other distributions for the nonzero elements of the interaction matrix give similar form for the fitness trajectory.
Fig. S8. Logarithmic fitness trajectories are also observed for different values of ρ.
Fig. S9. The decay of the two-time correlation function depends on both the time difference Δt and the initial time of the measurement tw, implying that the system ages.
Reference (45)
REFERENCES AND NOTES
- 1.Kryazhimskiy S., Tkačik G., Plotkin J. B., The dynamics of adaptation on correlated fitness landscapes. Proc. Natl. Acad. Sci. U.S.A. 106, 18638–18643 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wiser M. J., Ribeck N., Lenski R. E., Long-term dynamics of adaptation in asexual populations. Science 342, 1364–1367 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Frank S. A., Generative models versus underlying symmetries to explain biological pattern. J. Evol. Biol. 27, 1172–1178 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Jain K., Number of adaptive steps to a local fitness peak. Europhys. Lett. 96, 58006 (2011). [Google Scholar]
- 5.Neidhart J., Krug J., Adaptive walks and extreme value theory. Phys. Rev. Lett. 107, 178102 (2011). [DOI] [PubMed] [Google Scholar]
- 6.Schoustra S. E., Bataillon T., Gifford D. R., Kassen R., The properties of adaptive walks in evolving populations of fungus. PLOS Biol. 7, e1000250 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Szendro I. G., Schenk M. F., Franke J., Krug J., de Visser J. A. G. M., Quantitative analyses of empirical fitness landscapes. J. Stat. Mech. 2013, P01005 (2013). [Google Scholar]
- 8.Kouyos R. D., Leventhal G. E., Hinkley T., Haddad M., Whitcomb J. M., Petropoulos C. J., Bonhoeffer S., Exploring the complexity of the HIV-1 fitness landscape. PLOS Genet. 8, e1002551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salverda M. L. M., Dellus E., Gorter F. A., Debets A. J. M., van der Oost J., Hoekstra R. F., Tawfik D. S., de Visser J. A. G. M., Initial mutations direct alternative pathways of protein evolution. PLOS Genet. 7, e1001321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Blount Z. D., Borland C. Z., Lenski R. E., Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 105, 7899–7906 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinreich D. M., Chao L., Rapid evolutionary escape by large populations from local fitness peaks is likely in nature. Evolution 59, 1175–1182 (2005). [PubMed] [Google Scholar]
- 12.Weissman D. B., Desai M. M., Fisher D. S., Feldman M. W., The rate at which asexual populations cross fitness valleys. Theor. Popul. Biol. 75, 286–300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neher R. A., Shraiman B. I., Statistical genetics and evolution of quantitative traits. Rev. Mod. Phys. 83, 1283–1300 (2011). [Google Scholar]
- 14.Barrick J. E., Yu D. S., Yoon S. H., Jeong H., Oh T. K., Schneider D., Lenski R. E., Kim J. F., Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461, 1243–1247 (2009). [DOI] [PubMed] [Google Scholar]
- 15.Costanzo M., VanderSluis B., Koch E. N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S. D., Pelechano V., Styles E. B., Billmann M., van Leeuwen J., van Dyk N., Lin Z.-Y., Kuzmin E., Nelson J., Piotrowski J. S., Srikumar T., Bahr S., Chen Y., Deshpande R., Kurat C. F., Li S. C., Li Z., Usaj M. M., Okada H., Pascoe N., San Luis B.-J., Sharifpoor S., Shuteriqi E., Simpkins S. W., Snider J., Suresh H. G., Tan Y., Zhu H., Malod-Dognin N., Janjic V., Przulj N., Troyanskaya O. G., Stagljar I., Xia T., Ohya Y., Gingras A.-C., Raught B., Boutros M., Steinmetz L. M., Moore C. L., Rosebrock A. P., Caudy A. A., Myers C. L., Andrews B., Boone C., A global genetic interaction network maps a wiring diagram of cellular function. Science 353, aaf1420 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tong A. H. Y., Lesage G., Bader G. D., Ding H., Xu H., Xin X., Young J., Berriz G. F., Brost R. L., Chang M., Chen Y., Cheng X., Chua G., Friesen H., Goldberg D. S., Haynes J., Humphries C., He G., Hussein S., Ke L., Krogan N., Li Z., Levinson J. N., Lu H., Ménard P., Munyana C., Parsons A. B., Ryan O., Tonikian R., Roberts T., Sdicu A.-M., Shapiro J., Sheikh B., Suter B., Wong S. L., Zhang L. V., Zhu H., Burd C. G., Munro S., Sander C., Rine J., Greenblatt J., Peter M., Bretscher A., Bell G., Roth F. P., Brown G. W., Andrews B., Bussey H., Boone C., Global mapping of the yeast genetic interaction network. Science 303, 808–813 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Kauffman S. A., Weinberger E. D., The NK model of rugged fitness landscapes and its application to maturation of the immune response. J. Theor. Biol. 141, 211–245 (1989). [DOI] [PubMed] [Google Scholar]
- 18.Stadler P. F., Happel R., Random field models for fitness landscapes. J. Math. Biol. 38, 435–478 (1999). [Google Scholar]
- 19.Stadler P. F., Landscapes and their correlation functions. J. Math. Chem. 20, 1–45 (1996). [Google Scholar]
- 20.Dill K. A., Chan H. S., From Levinthal to pathways to funnels. Nat. Struct. Mol. Biol. 4, 10–19 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Onuchic J. N., Luthey-Schulten Z., Wolynes P. G., Theory of protein folding: The energy landscape perspective. Annu. Rev. Phys. Chem. 48, 545–600 (1997). [DOI] [PubMed] [Google Scholar]
- 22.Amit D. J., Gutfreund H., Sompolinsky H., Spin-glass models of neural networks. Phys. Rev. A 32, 1007–1018 (1985). [DOI] [PubMed] [Google Scholar]
- 23.Schneidman E., Berry M. J. II, Segev R., Bialek W., Weak pairwise correlations imply strongly correlated network states in a neural population. Nature 440, 1007–1012 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mani R., St. Onge R. P., Hartman J. L. IV, Giaever G., Roth F. P., Defining genetic interaction. Proc. Natl. Acad. Sci. U.S.A. 105, 3461–3466 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.M. A. Nowak, Evolutionary Dynamics (Harvard Univ. Press, 2006). [Google Scholar]
- 26.Iwasa Y., Michor F., Nowak M. A., Stochastic tunnels in evolutionary dynamics. Genetics 166, 1571–1579 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Good B. H., Desai M. M., The impact of macroscopic epistasis on long-term evolutionary dynamics. Genetics 199, 177–190 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.S. Gavrilets, Fitness Landscapes and the Origin of Species (MPB-41) (Princeton Univ. Press, 2004), vol. 41. [Google Scholar]
- 29.Van Nimwegen E., Crutchfield J. P., Optimizing epochal evolutionary search: Population-size dependent theory. Mach. Learn. 45, 77–114 (2001). [Google Scholar]
- 30.van Nimwegen E., Crutchfield J. P., Mitchell M., Finite populations induce metastability in evolutionary search. Phys. Lett. A 229, 144–150 (1997). [Google Scholar]
- 31.Bouchaud J.-P., Weak ergodicity breaking and aging in disordered systems. J. Physique I 2, 1705–1713 (1992). [Google Scholar]
- 32.M. Mézard, G. Parisi, M. Virasoro, Spin Glass Theory and Beyond: An Introduction to the Replica Method and Its Applications (World Scientific Publishing Company, 1987), vol. 9. [Google Scholar]
- 33.Mézard M., Parisi G., Virasoro M., Random free energies in spin glasses. J. Physique Lett. 46, 217 (1985). [Google Scholar]
- 34.Bray A. J., Moore M. A., Metastable states in spin glasses. J. Phys. C Solid State Phys. 13, L469–L476 (1980). [Google Scholar]
- 35.Khan A. I., Dinh D. M., Schneider D., Lenski R. E., Cooper T. F., Negative epistasis between beneficial mutations in an evolving bacterial population. Science 332, 1193–1196 (2011). [DOI] [PubMed] [Google Scholar]
- 36.Chou H.-H., Chiu H.-C., Delaney N. F., Segrè D., Marx C. J., Diminishing returns epistasis among beneficial mutations decelerates adaptation. Science 332, 1190–1192 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aguilar-Rodríguez J., Payne J. L., Wagner A., A thousand empirical adaptive landscapes and their navigability. Nat. Ecol. Evol. 1, 0045 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Amir A., Oreg Y., Imry Y., On relaxations and aging of various glasses. Proc. Natl. Acad. Sci. U.S.A. 109, 1850–1855 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.J.-P. Bouchaud, L. F. Cugliandolo, J. Kurchan, M. Mézard, in Spin Glasses and Random Fields, A. P. Young, Ed. (World Scientific, 1998), pp. 161–223. [Google Scholar]
- 40.Cugliandolo L. F., Kurchan J., On the out-of-equilibrium relaxation of the Sherrington-Kirkpatrick model. J. Phys. A Math. Gen. 27, 5749–5772 (1994). [Google Scholar]
- 41.Fukao K., Miyamoto Y., Slow dynamics near glass transitions in thin polymer films. Phys. Rev. E 64, 011803 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Nicodemi M., Coniglio A., Herrmann H. J., Frustration and slow dynamics of granular packings. Phys. Rev. E 55, 3962–3969 (1997). [Google Scholar]
- 43.Biroli G., Kurchan J., Metastable states in glassy systems. Phys. Rev. E 64, 016101 (2001). [DOI] [PubMed] [Google Scholar]
- 44.Kauffman S., Levin S., Towards a general theory of adaptive walks on rugged landscapes. J. Theor. Biol. 128, 11–45 (1987). [DOI] [PubMed] [Google Scholar]
- 45.Wahl L. M., Gerrish P. J., The probability that beneficial mutations are lost in populations with periodic bottlenecks. Evolution 55, 2606–2610 (2001). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/5/7/eaav3842/DC1
Section SA. Choice of null model for the combined effect of multiple independent mutations
Section SB. Range of validity for population size N
Section SC. Structural features of fitness landscapes
Section SD. Relaxation to local fitness maximum
Section SE. Hopping between MSs
Section SF. Batch culture
Fig. S1. Properties of fitness landscape as a function of L.
Fig. S2. Properties of fitness peaks as a function of L when additional weak interactions
between all sites are included.
Fig. S3. Figure showing how properties of MSs vary with fitness for ρ = 0.01 (black diamonds, Foffset = −3.4), ρ = 0.025 (red circles, Foffset = −3.8), ρ = 0.05 (green triangles, Foffset = −4.2), ρ = 0.075 (blue squares, Foffset = −4.4), and ρ = 0.1 (purple crosses, Foffset = −4.5).
Fig. S4. The number of connecting MSs ns, which is the number of MSs that the system can transit to next from the current MS, correlates with the number of double mutant escape paths out of a state np.
Fig. S5. Relaxation toward a single local fitness maximum slows down with increasing degree of epistasis.
Fig. S6. Changing the distribution of fixation probabilities does not significantly change the functional form of the fitness trajectory.
Fig. S7. Other distributions for the nonzero elements of the interaction matrix give similar form for the fitness trajectory.
Fig. S8. Logarithmic fitness trajectories are also observed for different values of ρ.
Fig. S9. The decay of the two-time correlation function depends on both the time difference Δt and the initial time of the measurement tw, implying that the system ages.
Reference (45)


