Abstract
In this study we interrogated the DNA methylome of myelofibrosis patients using high-density DNA methylation arrays. We detected 35,215 differentially methylated CpG, corresponding to 10,253 genes, between myelofibrosis patients and healthy controls. These changes were present both in primary and secondary myelofibrosis, which showed no differences between them. Remarkably, most differentially methylated CpG were located outside gene promoter regions and showed significant association with enhancer regions. This aberrant enhancer hypermethylation was negatively correlated with the expression of 27 genes in the myelofibrosis cohort. Of these, we focused on the ZFP36L1 gene and validated its decreased expression and enhancer DNA hypermethylation in an independent cohort of patients and myeloid cell-lines. In vitro reporter assay and 5’-azacitidine treatment confirmed the functional relevance of hyper-methylation of ZFP36L1 enhancer. Furthermore, in vitro rescue of ZFP36L1 expression had an impact on cell proliferation and induced apoptosis in SET-2 cell line indicating a possible role of ZFP36L1 as a tumor suppressor gene in myelofibrosis. Collectively, we describe the DNA methylation profile of myelofibrosis, identifying extensive changes in enhancer elements and revealing ZFP36L1 as a novel candidate tumor suppressor gene.
Introduction
Philadelphia chromosome-negative myeloproliferative neoplasms, namely polycythemia vera (PV), essential thrombocythemia (ET) and primary myelofibrosis (MF), are characterized by a clonal transformation of hematopoietic progenitors leading to expansion of fully differentiated myeloid cells.1 Primary MF carries the worst prognosis of all Philadelphia chromosome-negative myeloproliferative neoplasms, with progressive marrow fibrosis, extramedullary hematopoiesis, mild to severe splenomegaly and an increased risk of transformation into leukemia.2 Secondary MF can also arise from PV and ET (hereafter referred to as post-PV and post-ET MF, respectively) by mechanisms that are still poorly understood and are clinically and morphologically indistinguishable from primary MF.3
MF has been intensively studied from the genetic perspective;4,5 in fact, the modified World Health Organization (WHO) diagnostic criteria for Philadelphia chromosome-negative myeloproliferative neoplasms require the demonstration of a genetic marker of clonal hematopoiesis (JAK2V617F, CALR or MPL mutations).6 The frequency of mutations on relevant epigenetic genes (i.e., DNMT3A, EZH2 and ASXL1) suggests that MF might have an epigenetic component that, to our knowledge, remains poorly characterized.5 So far, epigenetic changes such as DNA methylation have been scarcely addressed in MF7 partly due to the limited changes in promoter DNA methylation compared to those in other hematologic malignancies, as previously published by our group.8 DNA methylation of CpG islands (CGI) (mostly on putative promoter regions) has been traditionally studied in both normal and neoplastic hematopoiesis.9,10 However, high-throughput platforms offer a wider coverage of the genome, allowing a better understanding of DNA methylation dynamics in regions distant from CGI.11 In this regard, enhancer regions have been characterized as potentially relevant sites of DNA methylation outside CGI.12–14 Chromatin immunoprecipitation-sequencing studies have enabled reliable mapping of genome-wide active enhancer regions based on histone modifications (e.g., H3K4me1 and H3K27Ac),15,16 allowing the identification of enhancers playing a role in dynamic transcriptional regulation during hematopoiesis.17
The present work describes a comprehensive genome-wide analysis of DNA methylation in MF patients, coupled with a gene expression analysis and information on functional chromatin states, compared with those of samples from healthy donors.16 Focusing on potential epigenetic alterations in enhancer regions, we identified ZFP36L1 as a potential tumor suppressor gene with relevance for the pathogenesis of MF.
Methods
Patients’ samples and clinical data
Samples from MF patients (n=39) were bone marrow, granulocytes or total peripheral blood cells. The MF cohort comprised cases of primary MF (n=22), post-ET MF (n=7) and post-PV MF (n=10). Peripheral blood cells from healthy donors (n=6) were used as control samples in this study. All patients were diagnosed using the 2008 version of the WHO classification system of hematologic malignancies.18 Data on JAK2V617F mutation status were retrospectively available for all patients, whereas no data on CALR and MPL mutations were available. The patients’ data are accessible from the Gene Expression Omnibus (GSE118241).
Samples and patients’ data were provided by the Biobank of the University of Navarra and were processed following standard operating procedures approved by the local Ethics & Scientific Committee. Prior to the collection of samples, all patients consented to the use of their data and to the use of stored material for research purposes.
DNA methylation profiling
DNA methylation was assessed using a Human-Methylation 450K Bead-Chip kit (Illumina, Inc., San Diego, CA, USA) and the data were analyzed by Bioconductor open source software. The analytical pipeline implemented several filters to exclude technical and biological biases and take into account the performance characteristics of Infinium I and Infinium II assays.19 Differentially methylated CpG were defined as previously described.13,19 Details on the experimental procedures, annotation of CpG sites, detection of differentially methylated regions, and Gene Ontology analysis20 are described in the Online Supplementary Methods.
Identification of candidate genes targeted by aberrant DNA methylation in enhancers
Data on gene expression profiling from primary MF and healthy peripheral blood samples were obtained from the publicly available Gene Expression Omnibus accession bank number GSE26049.21 Data were further processed using R and the open source Limma package.22 Further details are described in the Online Supplementary Methods.
Luciferase reporter assays
The CpG-free vector (pCPG-L), kindly provided by Dr. Michael Rehli,23 was used to clone the ZFP36L1 enhancer region. Luciferase experiments were performed in triplicate and the details are described in the Online Supplementary Methods. Primer sequences are available in Online Supplementary Table S1.
ZFP36L1 binding motif search
To further validate the potential relevance of the ZFP36L1 gene in MF, the DREME motif discovery algorithm24 was used to assess enrichment of genes with the ZFP36L1 consensus binding sequence among those genes differentially expressed in MF [false discovery rate (FDR)≤ 0.05].
Overexpression of ZFP36L1
A vector containing the ZFP36L1 open reading frame was kindly provided by Dr. Murphy and subcloned into a PL-SIN-GK vector.25 Further details are described in the Online Supplementary Methods.
Statistical analysis
For parametric group comparisons one-way analysis of variance (ANOVA) with the Dunnet correction was used, whereas for nonparametric group comparisons the Kruskall-Wallis test with the Dunn correction was employed. Paired data were analyzed with a Friedman non-parametric test with the Dunn correction for multiple comparisons, for the data with single measurements. Two-way ANOVA with the Tukey correction was used for data with multiple paired measurements. All tests were performed using Prism 7™ software (GraphPad, La Jolla, CA, USA).
Details of other experimental procedures are given in the Online Supplementary Methods.
Results
Myelofibrosis is characterized by a specific DNA methylation pattern enriched in enhancer regions
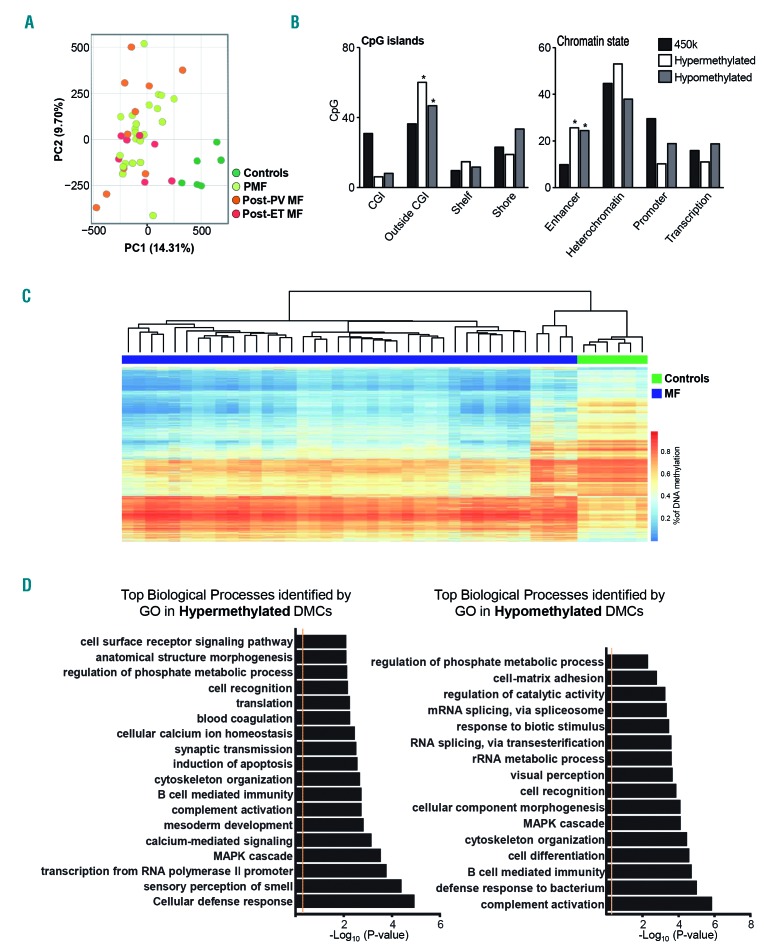
In order to provide an exhaustive analysis of the DNA methylation profile in patients with MF, we analyzed the DNA methylome of patients with primary MF, secondary MF (including post-ET/post-PV MF) and healthy donors as controls, using the Human- Methylation 450K array. The first result worth highlighting was the epigenetic similarity between primary and post-ET/post-PV MF. Interestingly, with a FDR<0.05, no differentially methylated CpGs were found between primary and secondary MF. Furthermore, we did not identify any differentially methylated CpG between post-ET and post-PV MF. However, both unsupervised principal component analysis (PCA) (Figure 1A) and hierarchical clustering studies (Online Supplementary Figure S1A) using all CpG analyzed confirmed an explicit segregation and a clear epigenetic difference between samples from patients with MF and those from healthy controls. These results allowed us hereafter to consider all MF samples as a single sample cohort.
Figure 1.
Patients with myelofibrosis have a different DNA methylation profile from controls, with changes located primarily in enhancer regions. (A) Unsupervised principal component analysis (PCA) showing a differential DNA methylation profile of myelofibrosis (MF) patients and healthy controls with no differences between primary and secondary MF. (B) Distribution of differentially methylated CpG according to CpG island mapping (left graph) or functional chromatin analysis (right graph) grouped by DNA methylation status of the probes (legend). *P≤0.05. (C) Heatmap of DNA methylation levels of differentially methylated CpG sites located in enhancer regions in MF patients and healthy controls. (D) GO-PANTHER analysis of genes adjacent to differenatially methylated CpG located in enhancer regions. Analysis of hypermethylated and hypomethylated genes is shown in the left and right panels, respectively. PC1: principal component 1; PC2: principal component 2; PMF: primary myelofibrosis; PV: polycythemia vera; MF: myelofibrosis; ET: essential thrombocythemia; CGI: CpG islands; DMC: differentially methylated CpG.
Next, we sought to interrogate differences in DNA methylation between MF samples and healthy controls. In this supervised analysis, we detected 35,215 differentially methylated CpG (FDR≤0.05) corresponding to 10,253 coding genes. Among all of these differentially methylated CpG, 65.3% were hypomethylated (corresponding to 22,998 CpG) and the remaining 34.7% were hypermethylated (a total of 12,217 CpG), suggesting that loss of DNA methylation is the predominant alteration in MF. Global DNA hypomethylation has also been a common finding in other hematologic malignancies such as chronic lymphocytic leukemia, multiple myeloma and acute myeloid leukemia.13,26,27
Analysis of the genomic location of differentially methylated CpG showed that both hyper- and hypomethylated CpG were underrepresented in classical CGI and significantly enriched outside CpG islands (Figure 1B). This is an interesting finding, because traditionally, neoplasms acquire hypomethylation outside CGI and hyper-methylation inside the islands,13,27 and suggests that patterns of methylation gain in MF might differ from those of other neoplasms. To shed light onto the specific function of the differentially methylated CpG, the chromatin state of each CpG was categorized adapting a publicly available annotation of chromatin immunoprecipitation-sequencing data from CD34+ hematopoietic progenitor cells, in which four distinct states were defined: promoter (with H3K4me3), active enhancer (with H3K4me1 and H3K27ac), transcribed regions (showing H3K36me3) and heterochromatin (including H3K9me3 and H3K27me3).16 Both hyper- and hypo-methylated CpG showed significant enrichment in enhancer regions, together with a striking underrepresentation in promoter regions (Figure 1B). Unsupervised clustering of differentially methylated CpG located exclusively in enhancer regions (Online Supplementary Table S2) displayed a clear segregation of the majority of MF patients from healthy controls (Figure 1C) identifying 4,182 hypermethylated and 10,935 hypomethylated probes. These results suggest that patients with MF show an intrinsic aberrant pattern of DNA methylation preferentially located in enhancer regions of the genome.
To further characterize the aberrant DNA methylation of enhancer regions in MF, GO-PANTHER enrichment analysis was performed separately in differentially methylated genes. GO terms with an adjusted FDR<0.05 were selected, showing in the case of hypermethylated enhancers relevant cellular processes such as cellular defense response or induction of apoptosis (Figure 1D).
DNA methylation of enhancer regions is associated with gene expression profile in myelofibrosis
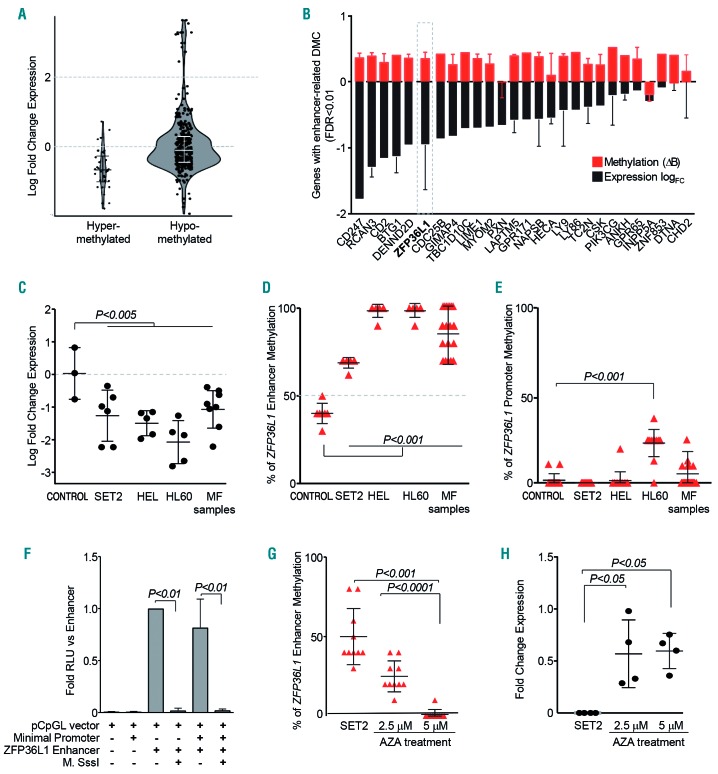
DNA methylation levels of enhancer regions were correlated with the expression of host and adjacent coding genes using publicly available gene expression data of an independent cohort of MF patients and healthy donors (GSE26049).21 Fold increases in gene expression values were grouped according to the hypermethylated (Δβ>0.4) or hypomethylated (Δβ<-0.4) enhancer status in MF versus controls. This analysis showed that enhancer DNA hyper-methylation was associated with decreased gene expression of host/adjacent coding genes. In contrast, hypomethylated enhancer regions were not associated with increased gene expression (Figure 2A).
Figure 2.
Aberrant enhancer DNA methylation regulates gene expression in myelofibrosis. (A) Violin density plots of expression of genes with differentially methylated CpG located in enhancer regions. The vertical axis represents log fold change in gene expression. The horizontal width of the plot represents density of data along the y axis. (B) Candidate genes with substantial changes in DNA methylation (FDR <0.01 and Δβ>0.4) and differential gene expression (logFC<0). Red bars represent the average DNA methylation of all enhancer-mapped probes, the black bars represent the average expression of all probes, and the error bars represent the standard deviation (SD) (C) ZFP36L1 downregulation validation by real-time quantitative polymerase chain reaction analysis of myelofibrosis (MF) patients and three myeloid cell lines (including SET-2) compared to healthy controls (n=3). (D,E) Bisulfite sequencing of the ZFP36L1 enhancer region (D) and promoter region (E) in healthy controls, cell lines and primary MF samples. For each sample, the graph shows the mean ± SD of ten CpG dinucleotides for enhancer regions and 15 CpG dinucleotides for promoter regions. (F) pCpG-L luciferase reporter assay showing the inhibition of luciferase activity after treatment of the ZFP36L1 enhancer region with Sss-I methyltransferase. (G) DNA methylation levels of the enhancer region – the same ten CpG dinucleotides as in (D) after 5-azacytidine treatment of SET-2. (H) ZFP36L1 expression levels after 5-azacytidine treatment of SET-2. Plots/bars indicate mean ± SD. FC: fold change; DMC: differentially methylated CpG; FDR: false discovery rate; CONTROL: healthy controls. MF; myelofibrosis; AZA: 5-azacytidine.
Next, we designed a more stringent approach to identify the set of genes underlying the most significant and substantial changes in enhancer DNA methylation (FDR <0.01, Δβ>0.4), coupled with downregulation of their expression (logFC<0) (Figure 2B). After identifying a number of potential candidates (27 genes), we focused on ZFP36L1, which codes for a RNA-binding protein that mediates the decay of unstable mRNA with AU rich elements in the 3′ untranslated region.28,29 Interestingly, the enhancer region associated with this candidate gene was located in its intragenic region, presumably acting as a cis-regulatory element of ZFP36L1 transcription. It is worth noting that this regulatory element was consistently hypermethylated in the cohort of MF patients and showed the largest number of hypermethylated enhancer-related CpG probes among the final 27-gene list.
ZFP36L1 enhancer hypermethylation correlated with downregulation of expression in MF as compared to controls (Figure 2B and Online Supplementary Figure S1B), which was further confirmed in an independent cohort of MF patients and myeloid cell lines (Figure 2C). Bisulfite sequencing confirmed that DNA methylation of the enhancer region of ZFP36L1 was consistently higher in all MF samples and myeloid cell lines than in control samples, whereas the promoter region remained unmethylated (Figure 2D,E and Online Supplementary Figure S1C). Results obtained from luciferase-reporting assays demonstrated that the exogenous DNA methylation significantly reduced ZFP36L1 enhancer activity (Figure 2F). Moreover, 5′-azacytidine hypomethylating treatment was able to reverse the DNA methylation levels of the enhancer region in vitro, partially restoring the gene expression levels of ZFP36L1 in the SET-2 cell line (Figure 2G,H).
ZFP36L1 acts as a tumor suppressor gene and potentially affects the myelofibrosis transcription profile
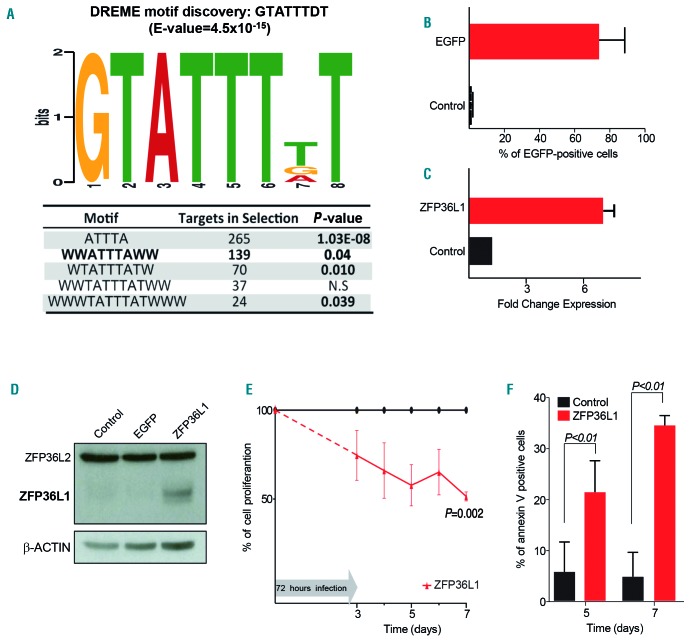
We hypothesized that ZFP36L1 downregulation could lead to upregulation of its putative targets in MF. We used DREME, a motif discovery algorithm specifically designed to find short, core DNA-binding motifs enriched in the 3′ untranslated region of genes. We found that the GTATTTDT motif (E-value=4.5×10−15) was in fact overrepresented in transcripts upregulated in MF patients (Figure 3A). Subsequently, an analysis of motif enrichment was performed, revealing a significant enrichment of upregulated genes in MF patients among the group containing the mentioned motif (P=7.69×10−20; logFC >1; P<0.05).
Figure 3.
ZFP36L1 rescue decreases cell viability in myelofibrosis. (A) Consensus binding motif for ZFP36L1 obtained by DREME motif discovery among transcripts with putative AU-rich motifs upregulated in myelofibrosis samples. (B) Efficiency of infection measured by the percentage of EGFP-positive cells after lentiviral infection. (C) Quantitative polymerase chain reaction validation of ZFP36L1 restoration in the SET-2 cell line after lentiviral infection. (D) ZFP36L1 protein restoration measured by western blot in the SET-2 cell line after lentiviral infection. (E,F) ZFP36L1 rescue with lentiviral vector infection in the SET-2 cell line decreased cell proliferation rate (E) and increased annexin V-positive cells (F).
To complement DREME analysis, we searched the AREsite30 database for AU-rich elements to determine whether we could detect, among the genes differentially expressed (B-value>10) between MF and controls, an enrichment of these sequences in the upregulated subset. Of all the possible AU motifs, we focused on the most restricted 9, 11 and 13-mer motifs. Interestingly, we were able to identify an enrichment of a 9-mer sequence WTATTTATW (P=0.01) and a 13-mer sequence WWW-TATTTATWWW (P=0.03) exclusively among the upregulated genes in MF patients (Figure 3A). Remarkably, both AU motifs strongly resemble the ZFP36L1 core-binding motif predicted by the DREME algorithm.
Re-expression of ZPF36L1 was achieved through lentiviral infection of the SET-2 cell line. Seventy-two hours after infection, the levels of EGFP-positive cells used as the positive control confirmed successful infection, and the level of expression of ZFP36L1 confirmed satisfactory overexpression of the gene (Figure 3B-D). Rescue of ZFP36L1 expression resulted in a decrease of more than 50% in cell proliferation, alongside an increase of annexin V-positive cells as measured by flow cell cytometry (Figure 3E,F).
Discussion
In the present study, we have extended previous knowledge regarding the DNA methylome in both primary and secondary MF, focusing specially on those CpG sites located in enhancer regions of the genome. A preliminary analysis of the global DNA methylome revealed the absence of DNA methylation differences between primary and secondary MF. This constitutes the first key finding of the present study and allowed us to use all the MF samples in a single cohort for further analysis. Primary and secondary MF are known to have very similar biological features, presenting symptoms and clinical course and in fact, both entities are treated indistinctively according to most published guidelines3,31 Nevertheless, some recent evidence from large retrospective trials has suggested that traditional prognostic factors may not be applicable to secondary MF as patients with post-ET MF seem to survive longer than those with post-PV MF or primary MF.3,31–33
The remarkably homogenous epigenetic profile of all our MF samples supports a common biological origin of primary and secondary MF.34 The DNA methylomes of the novel MF subtypes defined by the new 2016 WHO classification (prefibrotic and overt MF) remain to be characterized and it will be interesting to establish whether these subtypes have different methylation profiles. This aspect exceeded the possibilities of our cohort (retrospective availability of histology samples) but warrants further investigation.
Although previous studies have already interrogated the DNA methylation landscape of MF,7 their findings are limited to small numbers of epigenetic abnormalities mainly focused on the study of promoter regions. Our genome-wide approach of DNA methylation analysis using the 450k array allowed us to interrogate regulatory regions outside traditional promoters and obtain a deeper insight into the aberrant DNA methylome of MF. Changes in DNA methylation levels are known to cooperate with the deposition of chromatin marks, particularly H3K4 methylation, to render the enhancers/promoters accessible/inaccessible to the transcription machinery.35–37 Hence, the changes in DNA methylation observed in MF are expected to have an impact on the transcriptional profile of MF and potentially contribute to the MF malignant phenotype. Enhancer DNA methylation changes have been described to play a more prominent role in transcriptional regulation than promoter DNA methylation, governing processes such as hematopoietic differentiation and neoplastic transformation through the regulation of key transcription factors and genes.12,13,27,37,38 Translated into the context of Philadelphia chromosome-negative myeloproliferative neoplasms, this evidence might support the involvement of aberrant enhancer DNA methylation in the abnormal pattern of differentiation leading to MF. Enhancer hyper-methylation has been reported in neutrophils,12 B cells,39 AML cells26 and myeloma13 adding evidence to dynamic enhancer DNA hypermethylation as a relevant regulatory mechanism of gene expression both in normal and neoplastic hematopoietic cells.
Although the potential involvement of ZFP36L1 in myeloid differentiation has been described previously,40 our results suggest that epigenetic downregulation of ZFP36L1 might be a prominent event in the pathobiology of MF; more importantly, hypermethylation of an enhancer regulatory element represents a novel mechanism of disrupted gene expression in the context of MF and ZFP36L1. ZFP36L1 has been previously implicated in normal hematopoiesis41 and specifically associated with erythroid and myeloid differentiation,40 suggesting a possible role of this gene in MF onset and progression. Moreover, ZFP36L1 is also known to mediate mRNA decay of genes relevant to cell proliferation, survival and differentiation such as CDK6, TNFα, BCL2, NOTCH1 and STAT5B.42,43 Interestingly, the enhancer region associated with this candidate gene was consistently hypermethylated in the cohort of MF patients and was located in its intragenic region, presumably acting as a cis-regulatory element of ZFP36L1 transcription. The motif discovery experiments support our hypothesis of epigenetic deregulation of ZFP36L1, suggesting that MF samples with ZFP36L1 loss of expression experience upregulation of the gene’s putative targets. Consequently, when ZFP36L1 expression levels are restored with the lentiviral model, SET-2 cells lose their malignant proliferative phenotype, strengthening the tumor suppressor role of this gene in MF. Taken together, these results link ZFP36L1 to the pathobiology of MF, ultimately resulting in transcriptome deregulation of genes relevant to cell proliferation, survival and differentiation, as previously described.40,42,44,45
Conclusion
The DNA methylation landscape of patients with primary MF or post-ET/post-PV MF is consistently different from that of healthy individuals. The absence of differences between primary MF and post-ET/post-PV MF suggests that the changes seen in MF are founding epigenetic alterations occurring at the level of stem cells of this myeloproliferative neoplasm and maintained in differentiated myeloid cells. Aberrant DNA methylation in MF is predominantly located in enhancer regions and has a significant impact on the expression of their target genes. Combining DNA methylation and gene expression data, we identified ZFP36L1 as an attractive new possible therapeutic target that shows a decrease of gene expression mediated by enhancer hypermethylation. Our results also suggest a direct effect of ZFP36L1 downregulation on the gene expression profile of MF, through upregulation of mRNA harboring canonical sites with AU-rich elements. In vitro rescue of ZFP36L1 expression had an impact on cell proliferation and induced apoptosis in the SET-2 cell line, indicating a possible role of ZFP36L1 as a tumor suppressor gene in MF. Moreover, treatment with 5’-azacytidine further evidenced the plausibility of ZFP36L1 pharmacological manipulation. Taken together, these results provide evidence of an unexplored therapeutic target in MF patients, which remains to be properly evaluated in the pre-clinical setting.
Acknowledgments
We particularly acknowledge the patients for their participation and the Biobank of the University of Navarra for its collaboration. We thank John J Murphy and Amor Alcaraz for providing ZFP36L1 expression constructs. This research was funded by grants from Instituto de Salud Carlos III (ISCIII) PI14/01867, PI16/02024 and PI17/00701, TRASCAN (EPICA), CIBERONC (CB16/12/00489; co-financed with FEDER funds), RTICC (RD12/0036/0068) and the Departamento de Salud del Gobierno de Navarra 40/2016. NM is supported by a FEHH-Celgene research grant, MP was supported by a Sara Borrell fellowship CD12/00540 and RO was supported by the Ministerio de Ciencia, Innovación y Universidades of Spain, Subprograma de Formación de Profesorado Universitario (FPU) award number FPU14/04331.
Footnotes
Check the online version for the most updated information on this article, online supplements, and information on authorship & disclosures: www.haematologica.org/content/104/8/1572
References
- 1.Spivak JL. Myeloproliferative neoplasms. N Engl J Med. 2017;376(22):2168–2181. [DOI] [PubMed] [Google Scholar]
- 2.Kim J, Haddad RY, Atallah E. Myeloproliferative neoplasms. Dis Mon. 2012;58(4):177–194. [DOI] [PubMed] [Google Scholar]
- 3.Passamonti F, Rumi E, Caramella M, et al. A dynamic prognostic model to predict survival in post-polycythemia vera myelofibrosis. Blood. 2008;111(7):3383–3387. [DOI] [PubMed] [Google Scholar]
- 4.Kim SY, Im K, Park SN, Kwon J, Kim J-A, Lee DS. CALR, JAK2, and MPL mutation profiles in patients with four different subtypes of myeloproliferative neoplasms: primary myelofibrosis, essential thrombocythemia, polycythemia vera, and myeloproliferative neoplasm, unclassifiable. Am J Clin Pathol. 2015;143(5):635–644. [DOI] [PubMed] [Google Scholar]
- 5.Vainchenker W, Kralovics R. Genetic basis and molecular pathophysiology of classical myeloproliferative neoplasms. Blood. 2017;129(6):667–679. [DOI] [PubMed] [Google Scholar]
- 6.Arber DA, Orazi A, Hasserjian R, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127(20):2391–2405. [DOI] [PubMed] [Google Scholar]
- 7.Myrtue Nielsen H, Lykkegaard Andersen C, Westman M, et al. Epigenetic changes in myelofibrosis: distinct methylation changes in the myeloid compartments and in cases with ASXL1 mutations. Sci Rep. 2017;7(1):6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pérez C, Pascual M, Martín-Subero JI, et al. Aberrant DNA methylation profile of chronic and transformed classic Philadelphia-negative myeloproliferative neoplasms. Haematologica. 2013;98(9):1414–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mendizabal I, Yi SV. Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation. Hum Mol Genet. 2016;25(1):69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deaton AM, Webb S, Kerr ARW, et al. Cell type-specific DNA methylation at intragenic CpG islands in the immune system. Genome Res. 2011;21(7):1074–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sandoval J, Heyn H, Moran S, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692–702. [DOI] [PubMed] [Google Scholar]
- 12.Rönnerblad M, Andersson R, Olofsson T, et al. Analysis of the DNA methylome and transcriptome in granulopoiesis reveals timed changes and dynamic enhancer methylation. Blood. 2014;123(17):e79–e89. [DOI] [PubMed] [Google Scholar]
- 13.Agirre X, Castellano G, Pascual M, et al. Whole-epigenome analysis in multiple myeloma reveals DNA hypermethylation of B cell-specific enhancers. Genome Res. 2015;25(4):478–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bell RE, Golan T, Sheinboim D, et al. Enhancer methylation dynamics contribute to cancer plasticity and patient mortality. Genome Res. 2016;26(5):601–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst J, Kheradpour P, Mikkelsen TS, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473(7345):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wijetunga NA, Delahaye F, Zhao YM, et al. The meta-epigenomic structure of purified human stem cell populations is defined at cis-regulatory sequences. Nat Commun. 2014;5:5195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lara-Astiaso D, Weiner A, Lorenzo-Vivas E, et al. Immunogenetics. Chromatin state dynamics during blood formation. Science. 2014;345(6199):943–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tefferi A, Thiele J, Vardiman JW. The 2008 World Health Organization classification system for myeloproliferative neoplasms. Cancer. 2009;115(17):3842–3847. [DOI] [PubMed] [Google Scholar]
- 19.Kulis M, Merkel A, Heath S, et al. Whole-genome fingerprint of the DNA methylome during human B cell differentiation. Nat Genet. 2015;47(7):746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mi H, Huang X, Muruganujan A, et al. PANTHER version 11: expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skov V, Larsen TS, Thomassen M, et al. Whole-blood transcriptional profiling of interferon-inducible genes identifies highly upregulated IFI27 in primary myelofibrosis. Eur J Haematol. 2011;87(1):54–60. [DOI] [PubMed] [Google Scholar]
- 22.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klug M, Rehli M. Functional analysis of promoter CpG methylation using a CpG-free luciferase reporter vector. Epigenetics. 2006;1(3):127–130. [DOI] [PubMed] [Google Scholar]
- 24.Bailey TL. DREME: motif discovery in transcription factor ChIP-seq data. Bioinformatics. 2011;27(12):1653–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hotta A, Cheung AYL, Farra N, et al. Isolation of human iPS cells using EOS lentiviral vectors to select for pluripotency. Nat Methods. 2009;6(5):370–376. [DOI] [PubMed] [Google Scholar]
- 26.Qu Y, Siggens L, Cordeddu L, et al. Cancer-specific changes in DNA methylation reveal aberrant silencing and activation of enhancers in leukemia. Blood. 2017;129(7): e13–e25. [DOI] [PubMed] [Google Scholar]
- 27.Kulis M, Heath S, Bibikova M, et al. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44(11):1236–1242. [DOI] [PubMed] [Google Scholar]
- 28.Hitti E, Bakheet T, Al-Souhibani N, et al. Systematic analysis of AU-rich element expression in cancer reveals common functional clusters regulated by key RNA-binding proteins. Cancer Res. May 2016. [DOI] [PubMed] [Google Scholar]
- 29.Baou M, Norton JD, Murphy JJ. AU-rich RNA binding proteins in hematopoiesis and leukemogenesis. Blood. 2011;118(22):5732–5740. [DOI] [PubMed] [Google Scholar]
- 30.Gruber AR, Fallmann J, Kratochvill F, Kovarik P, Hofacker IL. AREsite: a database for the comprehensive investigation of AU-rich elements. Nucleic Acids Res. 2011;39(Database issue):D66–D69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reilly JT, McMullin MF, Beer PA, et al. Use of JAK inhibitors in the management of myelofibrosis: a revision of the British Committee for Standards in Haematology Guidelines for Investigation and Management of Myelofibrosis 2012. Br J Haematol. 2014;167(3):418–420. [DOI] [PubMed] [Google Scholar]
- 32.Hernández-Boluda J-C, Pereira A, Gómez M, et al. The International Prognostic Scoring System does not accurately discriminate different risk categories in patients with post-essential thrombocythemia and post-polycythemia vera myelofibrosis. Haematologica. 2014;99(4):e55–e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Passamonti F, Giorgino T, Mora B, et al. A clinical-molecular prognostic model to predict survival in patients with post polycythemia vera and post essential thrombocythemia myelofibrosis. Leukemia. 2017;31(12):2726–2731. [DOI] [PubMed] [Google Scholar]
- 34.Brecqueville M, Rey J, Devillier R, et al. Array comparative genomic hybridization and sequencing of 23 genes in 80 patients with myelofibrosis at chronic or acute phase. Haematologica. 2014;99(1):37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharifi-Zarchi A, Gerovska D, Adachi K, et al. DNA methylation regulates discrimination of enhancers from promoters through a H3K4me1-H3K4me3 seesaw mechanism. BMC Genomics. 2017;18(1):964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.King AD, Huang K, Rubbi L, et al. Reversible regulation of promoter and enhancer his-tone landscape by DNA methylation in mouse embryonic stem cells. Cell Rep. 2016;17(1):289–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blattler A, Yao L, Witt H, et al. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15(9):469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hon GC, Rajagopal N, Shen Y, et al. Epigenetic memory at embryonic enhancers identified in DNA methylation maps from adult mouse tissues. Nat Genet. 2013;45(10):1198–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bergmann AK, Castellano G, Alten J, et al. DNA methylation profiling of pediatric B-cell lymphoblastic leukemia with KMT2A rearrangement identifies hypomethylation at enhancer sites. Pediatr Blood Cancer. 2017;64(3):e26251. [DOI] [PubMed] [Google Scholar]
- 40.Vignudelli T, Selmi T, Martello A, et al. ZFP36L1 negatively regulates erythroid differentiation of CD34+ hematopoietic stem cells by interfering with the Stat5b pathway. Mol Biol Cell. 2010;21(19):3340–3351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Galloway A, Saveliev A, Łukasiak S, et al. RNA-binding proteins ZFP36L1 and ZFP36L2 promote cell quiescence. Science. 2016;352(6284):453–459. [DOI] [PubMed] [Google Scholar]
- 42.Zekavati A, Nasir A, Alcaraz A, et al. Post-transcriptional regulation of BCL2 mRNA by the RNA-binding protein ZFP36L1 in malignant B cells. PLoS ONE. 2014;9(7): e102625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu J, Lu W, Liu S, et al. ZFP36L2, a novel AML1 target gene, induces AML cells apoptosis and inhibits cell proliferation. Leuk Res. 2018;68:15–21. [DOI] [PubMed] [Google Scholar]
- 44.Chen M-T, Dong L, Zhang X-H, et al. ZFP36L1 promotes monocyte/macrophage differentiation by repressing CDK6. Sci Rep. 2015;5(1):16229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hodson DJ, Janas ML, Galloway A, et al. Deletion of the RNA-binding proteins ZFP36L1 and ZFP36L2 leads to perturbed thymic development and T lymphoblastic leukemia. Nat Immunol. 2010;11(8):717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]