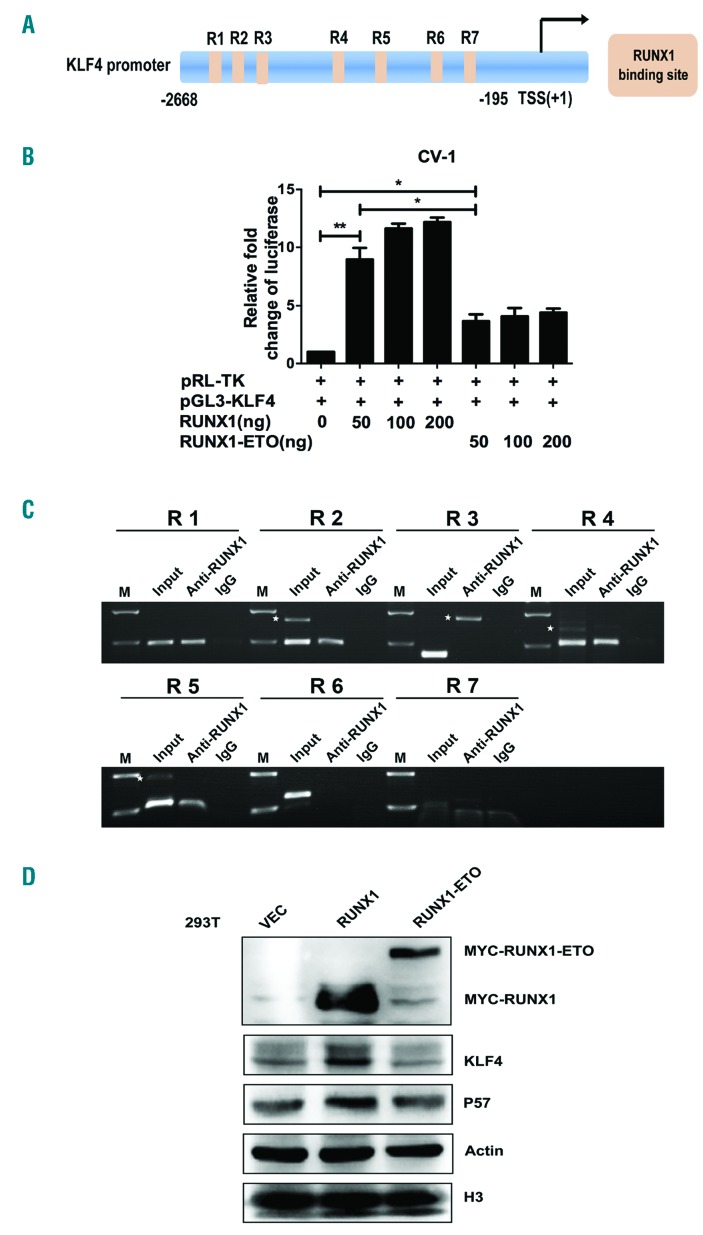
Figure 2.
KLF4 is a novel target gene of RUNX1 and RUNX1-ETO. (A) Schematic representation of KLF4 promoter fragments fused to a pGL3-Basic Vector. The putative RUNX1 binding sites are indicated by orange boxes as follows: R1 (-2617 to -2607), R2 (-2312 to -2302), R3 (-2014 to -2004), R4 (-1136 to -1126), R5 (-764 to -754), R6 (-399 to -394), and R7 (-263 to -258). Transcription start site (TSS) is indicated by an arrow. Numbers represent base pairs relative to TSS. (B) pGL3-KLF4 promoter reporter plasmid (pGL3-KLF4) was co-transfected with increasing doses of pCMV5-RUNX1 or pCMV5-RUNX1-ETO into CV-1 cells. At 48 h after transfection, the luciferase transcriptional activity of pGL3-KLF4 was measured and normalized to that of Renilla luciferase. (C) Chromatin immunoprecipitation analysis of RUNX1 binding sites to KLF4 promoter region in Kasumi-1 cells. The predicted binding sites R1, R2, R4 and R5 were successfully amplified, both from the input DNA and chromatin immunoprecipitation by an anti-RUNX1 antibody, whereas no amplified product was obtained in the IgG control group. White stars indicate non-specific amplified bandings. (D) 293T cells were transfected with pCMV5-vector, pCMV5-RUNX1 and pCMV5-RUNX1-ETO, respectively. At 48 h after transfection, cells were harvested and the protein levels of KLF4 were assayed by western blot. β-actin and H3 were used as protein loading controls.