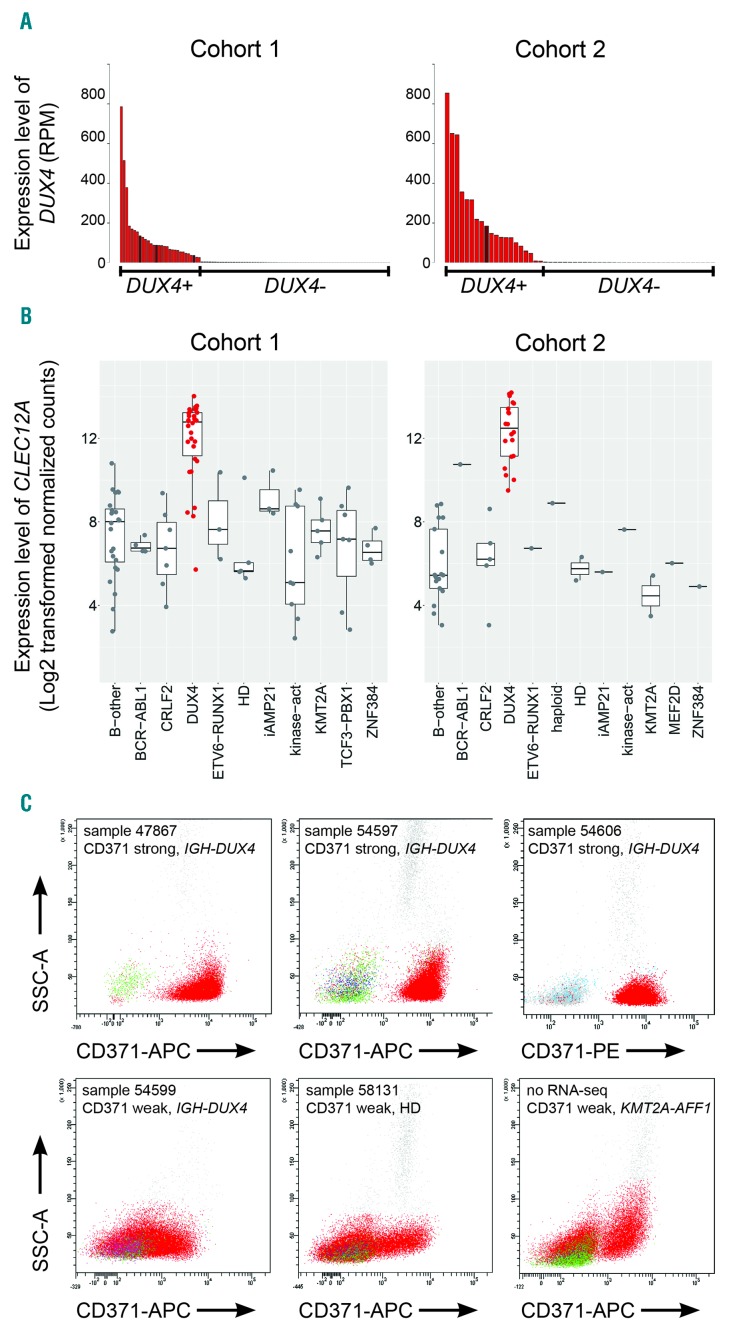
Figure 1.
Identification of DUX4-positive leukemia. (A) The number of reads that mapped to DUX4 cDNA (NM_001293798.2) per million total mapped reads assigned to RefSeq entries (RPM) for cohort 1 (left panel) and cohort 2 (right panel) are shown. DUX4+ cases are represented in red; cases with elevated DUX4 expression but lacking detectable IGH-DUX4 fusion transcripts are represented in brown. (B) Boxplots showing the expression levels of CLEC12A depicted as log2-transformed normalized counts calculated by DESeq2 in cohort 1 (101 samples from 92 patients) and cohort 2 (55 patients) (left and right panel, respectively). (C) Representative FACS plots of primary bone marrow cells from patients with B-cell acute lymphoblastic leukemia (B-ALL) stained for CD371-APC or CD371-PE; blast cells are depicted in red. B-other, B-ALL cases lacking any sentinel alteration; CRLF2, cases with P2YR8-CRLF2 or IGH-CRLF2 rearrangement; DUX4, cases with expression of DUX4; haploid, masked near haploid; HD, high hyperdiploid; iAMP21, intrachromosomal amplification of chromosome 21; kinase-act, cases harboring a kinase activation fusion gene; KMT2A, KMT2A fusion gene; MEF2D, MEF2D fusion gene; ZNF384, ZNF384 fusion gene; SSC: side scatter; APC: allophycocyanin; PE: phycoerythrin; RNA-seq: RNA-sequencing.