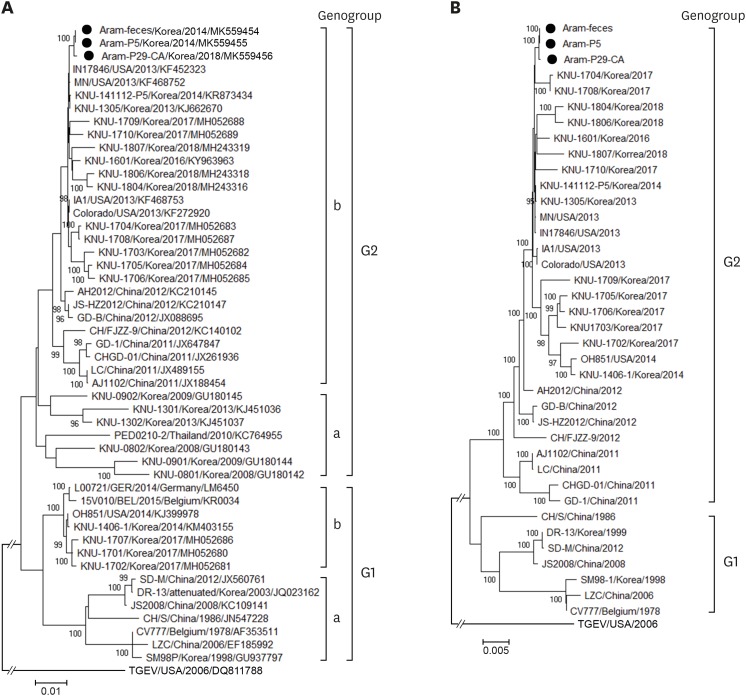
Fig. 3.
Phylogenetic analyses based on the nucleotide sequences of the spike (S) genes (A) and full-length genomes (B) of the porcine epidemic diarrhea virus strains. A region of the S gene and the complete genome sequence of transmissible gastroenteritis virus were included as the outgroups in each tree. Multiple sequence alignments were performed using ClustalX software and phylogenetic trees were constructed from the aligned nucleotide sequences using the neighbor-joining method. Numbers at each branch are bootstrap values greater than 50% based on 1000 replicates. The names of the strains, countries and dates (year) of isolation, GenBank accession numbers, and genogroups and subgroups are shown. Solid circles indicate the Aram strains described in this study. Scale bars indicate nucleotide substitutions per site.