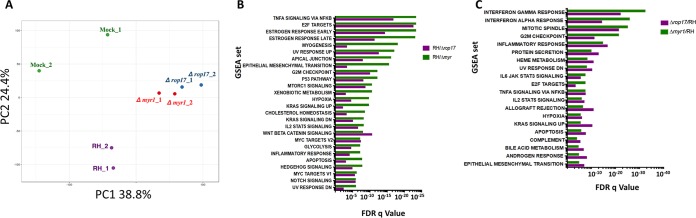
FIG 7.
Disrupting MYR1 or ROP17 has congruent impacts on the infected host cell’s transcriptome as assessed by RNASeq. (A) Principal-component analysis (PCA) of the RPKM values of HFFs mock infected or infected with RH-WT (wild type), RHΔmyr1, or RHΔrop17 tachyzoites. On a plot of PC1 and PC2, there is close similarity of the data for cells infected with the two mutants, RHΔmyr1 and RHΔrop17, relative to the mock- or RH-WT-infected cells. Gene names and RPKMs can be found in Table S1 in the supplemental material. (B) Gene set expression analysis (GSEA) of all genes expressed 2.5-fold higher in RH than in RHΔmyr1 (green) or RHΔrop17 (purple), where either was lower than the FDR q-value threshold of 10−5. The gene sets are ordered based on descending levels of significance (ascending q-values) for the cells infected with RHΔmyr1. (C) As in panel B, except GSEA was performed on the genes that were expressed 2.5-fold lower in RH compared to RHΔmyr1 and RHΔrop17 by the same criteria.