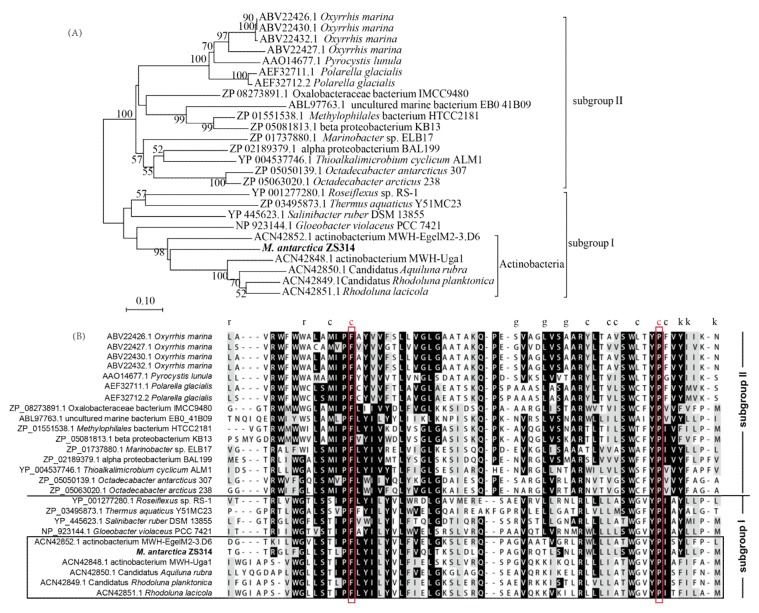
Figure 5.
Comparison of xanthorhodopsins from M. antarctica ZS314T and other representative microbial groups used in a previous study [27]. (A) Neighbor-joining tree of xanthorhodopsins based on amino acid sequences. Bootstrap values below 50 were not shown. (B) Multiple alignments of the putative keto-carotenoid-binding region of the xanthorhodopsins in (A), as identified previously [27,28]. Same as in the previous study [27], residues are marked by the letters r, c, g, and k to indicate contact with the ring, chain, glucoside and keto group of the carotenoid. The alignment was made using the ClustalW program online (https://www.genome.jp/tools-bin/clustalw) and shaded by the BoxShade program (https://embnet.vital-it.ch/software/BOX_form.html). Similar and identical residues in the alignment are highlighted with grey and black colors on the background, respectively.