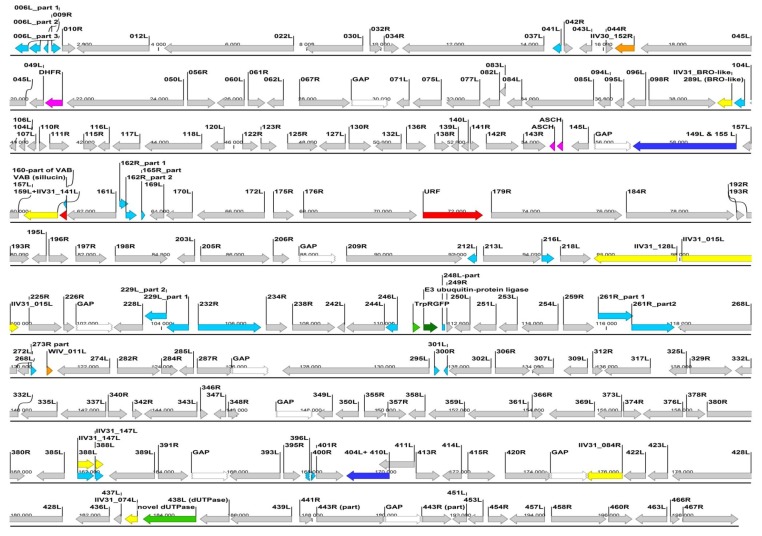
Figure 3.
Putative genomic map of the chameleon IIV isolate (Ir. iso. 1) constructed based on nine contigs of an Illumina de novo assembly, and applying the ORF numbering of IIV6. Red dotted rectangles indicate the genome regions, which were earlier covered by a Sanger method based comparison of the isolates (please note that the gaps joining the contigs are not proportional to their actual probable size, but were arbitrarily set to 1 kbp).  ORF highly similiar (85%–100%) to IIV6 homolog.
ORF highly similiar (85%–100%) to IIV6 homolog.  ORF broken or much shorter (>20%) than IIV6 homolog.
ORF broken or much shorter (>20%) than IIV6 homolog.  ORF is joined into one ORF from two IIV6 homologs.
ORF is joined into one ORF from two IIV6 homologs.  ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.
ORF most similar to an IIV31 (genus Iridovirus) gene/ORF.  ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.
ORF most similar to a polyiridovirus (genus Chlorididovirus) gene.  ORF most similar to a gene from a non-IV large DNA virus.
ORF most similar to a gene from a non-IV large DNA virus.  ORF most similar to a gene from a eukaryotic organism.
ORF most similar to a gene from a eukaryotic organism.  ORF most similar to a gene from a prokaryote.
ORF most similar to a gene from a prokaryote.  ORF with no homology to current GenBank entries.
ORF with no homology to current GenBank entries.  GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).
GAP (not yet sequenced part, arbitraty 1000, N’s joining of the mapped contigs).