Figure EV2. Upon stress, inhibition of translation prevents the recruitment of HSPA1A and HSPA8 inside nucleoli (related to Fig 2).
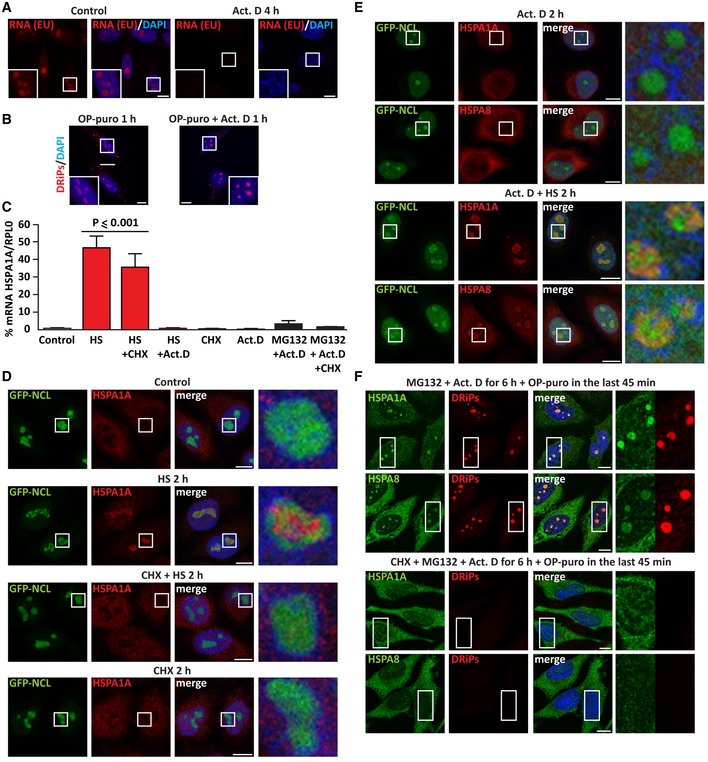
- Staining of newly synthesized RNA with 5‐ethynyl uridine (EU, 200 μM) alone or with actinomycin D (Act. D; 4 μM) for 4 h, followed by click chemistry. Nucleic acid is stained with DAPI.
- DRiP labeling in HeLa cells treated with OP‐puro (25 μM) alone or combined with Act. D (4 μM) for 1 h.
- HeLa cells were either left untreated (Control) or treated as follows: HS at 42°C for 3 h (HS), alone or combined with CHX (50 μg/ml) or Act. D (3 μM); single CHX or Act. D treatment for 3 h; transcriptional stress for 6 h (MG132 10 μM with Act. D 4 μM) (Audas et al, 2016), alone or combined with CHX (50 μg/ml). After cellular treatment, total RNA was extracted and mRNA levels of HSPA1A were measured by RT–qPCR (RPL0 was used for normalization). Statistical significance via one‐way ANOVA; n = 3, ± s.e.m.
- GFP‐NCL HeLa Kyoto cells were left untreated or exposed to HS at 42°C for 2 h alone or with CHX; as control, cells were also treated with CHX alone for 2 h (50 μg/ml). Cells were then fixed and stained for HSPA1A and DAPI.
- Staining of HSPA1A and HSPA8 in GFP‐NCL HeLa Kyoto cells treated with Act. D (3 μM) alone or combined with HS at 42°C for 2 h.
- Staining of HSPA1A, HSPA8, and DRiPs in HeLa cells subjected to transcriptional stress (MG132 10 μM with Act. D 4 μM) alone or concomitant to translation inhibition (CHX 50 μg/ml) for 6 h. OP‐puro (25 μM) was added during the last 45 min of treatment.
