-
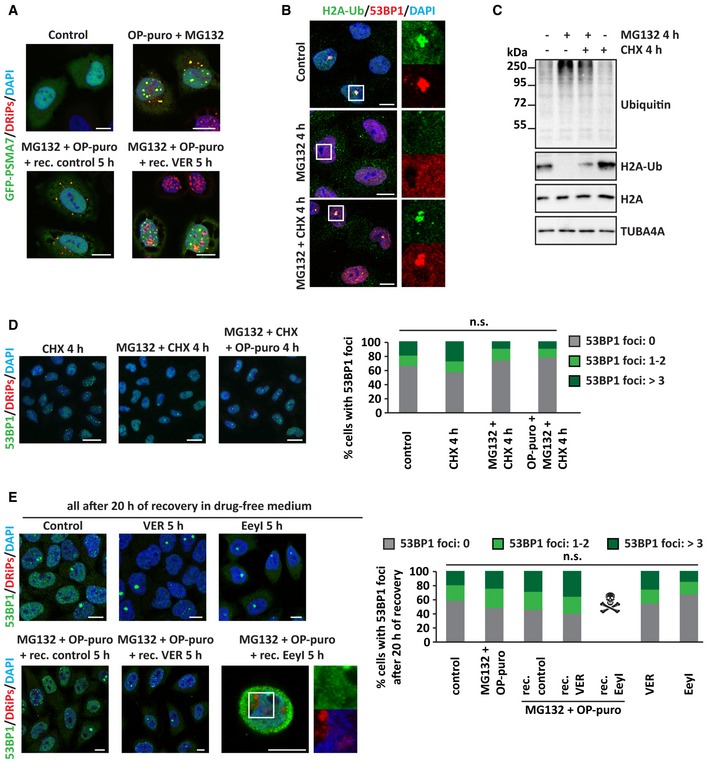
A
GFP‐PSMA7 HeLa Kyoto cells were left untreated or treated as described. Cells were fixed and subjected to click chemistry to visualize DRiPs. Nucleic acid was stained with DAPI. Scale bars: 10 μm.
-
B, C
Hela cells were left untreated or treated for 4 h with MG132 (10 μM), alone or combined with CHX (50 μg/ml). (B) Cells were fixed and processed for immunostaining of H2A‐Ub and 53BP1. Scale bars: 10 μm. (C) Expression levels of ubiquitin, H2A‐Ub, and H2A in total protein extracts. TUBA4A was used as loading control.
-
D
HeLa cells were left untreated or treated for 4 h with CHX, MG132, OP‐puro, combined as indicated and using the concentrations previously reported. Representative pictures of 53BP1 distribution and quantitation of the % of cells with 53BP1 foci/nucleus are shown. Number of cells counted/condition: 681–1,214 in three independent experiments; statistical significance via one‐way ANOVA; P = not significant (n.s.). Scale bars: 20 μm.
-
E
Cells treated as described, using the concentrations previously reported, were allowed to recover in drug‐free medium for 20 h. Cells were then fixed, and 53BP1 and DRiPs were stained. Quantitation of the % of cells with 53BP1 foci/nucleus is shown. Number of cells counted/condition: 668–1,678 in three independent experiments; statistical significance via one‐way ANOVA; P = not significant (n.s.). Scale bars: 10 μm.