Figure 1. ESCRT‐independent functions of the Dsc complex in membrane proteostasis.
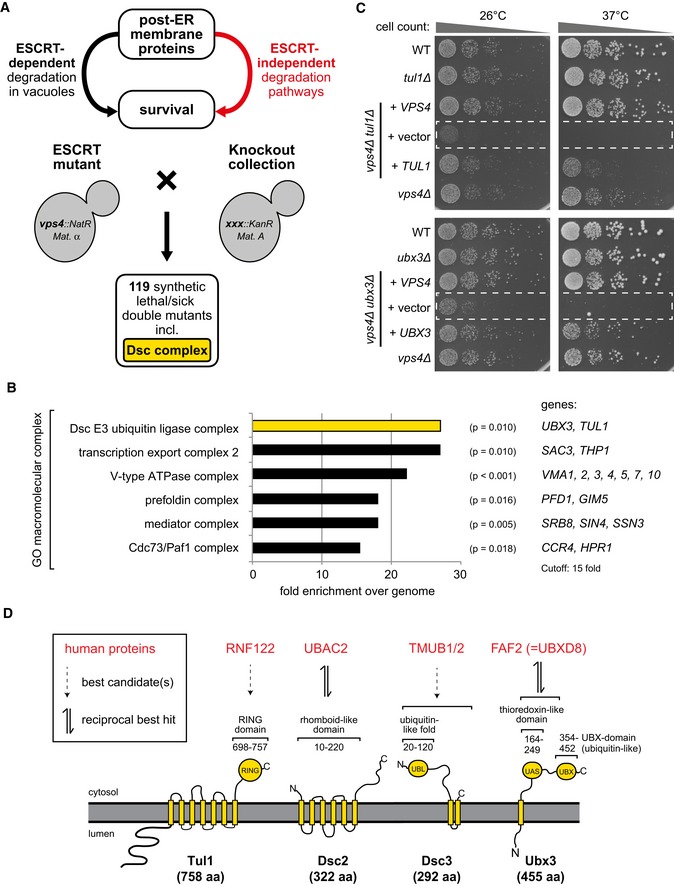
- Synthetic genetic array technology of Saccharomyces cerevisiae was used to cross the ESCRT mutant (query mutation vps4::NatR) with all non‐essential genes (xxx::KanR). The growth of double mutants (vps4::NatR xxx::KanR) was scored in two independent replicates for normal growth, synthetic sick, or synthetic lethal phenotypes. 119 genes (Appendix Table S1) showed synthetic sickness in one replicate and synthetic lethality in the other replicate or synthetic lethality in both replicates.
- Gene Ontology (GO) analysis for macromolecular complexes of the 119 genes that showed synthetic sickness in one replicate and synthetic lethality in the other replicate or synthetic lethality in both replicates (Appendix Tables S1 and S2). A hypergeometric test was used to estimate whether the mapped GO term is significantly enriched with the selected genes. Significant GO terms (P < 0.05) and their associated genes identified in the screen are shown with a cutoff for 15‐fold enrichment over the genome frequency.
- Equal amounts of WT cells and indicated single or double mutants in serial dilutions were incubated on agar plates at the indicated temperatures. The synthetic growth defects of vps4∆ tul1∆ or vps4∆ ubx3∆ double mutants at 26 and 37°C were complemented with plasmids encoding VPS4 or TUL1 or UBX3. The dashed boxes indicate the non‐complemented double mutants.
- Cartoon presentation of Dsc complex members and evolutionary conserved functional domains with human candidate genes that show the highest homology in the indicated domains.
