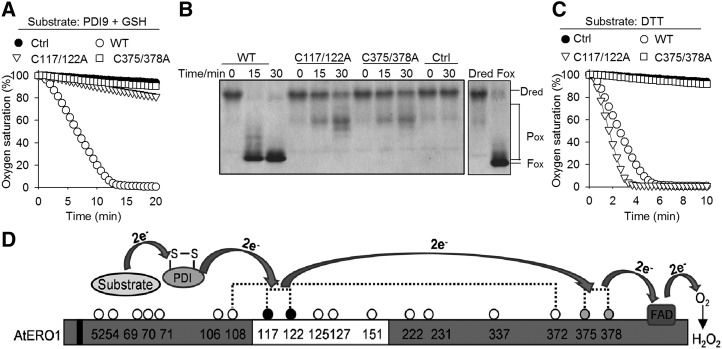
Figure 6.
Outer active site and inner active site are required for AtERO1 function. A, Oxygen consumption assay of AtERO1 mutants using GSH as a substrate in the AtERO1-AtPDI9 system. The reaction contains AtERO1/2, FAD, AtPDI9, and GSH. Ctrl, Reaction without AtERO1; WT, wild type. B, Analysis of activities of AtERO1 mutants using gel-based RNase A reoxidation assay in the presence of AtPDI9. Dred RNase A and Fox RNase A are shown at far right. C, Analysis of activities of AtERO1 mutants using DTT as a substrate. The reaction contains AtERO1/2, FAD, and DTT. D, Schematic representation of the electron transfer from the substrate via AtERO1 to oxygen in the presence of PDI. The electrons are transferred from AtPDIs to the inner active site of AtERO1 via the outer active site. The buried inner active site is oxidized by FAD, which reduces molecular oxygen, generating peroxide. The white box indicates the putative flexible loop region. Dotted lines indicate the putative disulfide bonds in AtERO1 based on the amino acid sequence alignment of Ero1 homologs. The Cys residues are represented by circles with numbering. The gray and black circles indicate the inner active site and the outer active site, respectively. The black box indicates the transmembrane region of AtERO1.