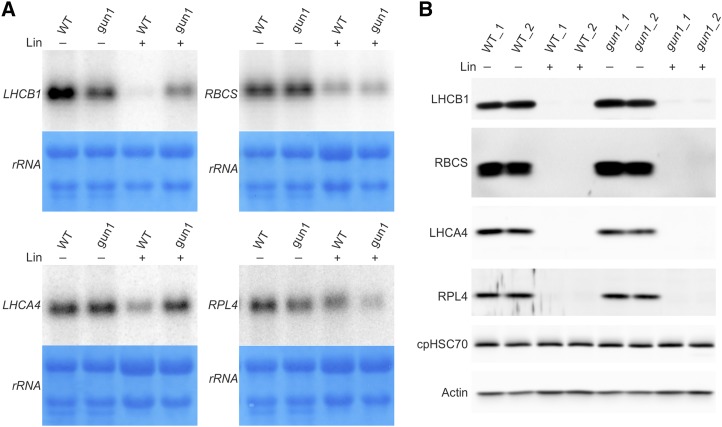
Figure 4.
Northern-blot and western-blot analyses to reveal the regulation of different types of HiToP genes. A, Northern-blot analysis of selected HiToP genes. Unlike LHCB1 (the probe used does not distinguish between different isoforms of LHCB1), whose transcript accumulation is largely repressed by Lin treatment in the wild type (WT), the transcripts of LHCA4 and RBCS (the probe used does not distinguish between different isoforms of RBCS) are less repressed and the RPL4 transcripts are nearly unchanged. Seeds from the wild type and gun1 were germinated and grown for 5 d in the dark followed by 2 d in continuous light in the absence (−) or presence (+) of 0.5 mm Lin. Methylene Blue staining of rRNAs after blotting was performed as a loading control. B, At the protein level, LHCA4, RBCS, and RPL4 are all below the detection limit of immunoblots both in the wild type and gun1 under Lin treatment. For LHCB1 in Lin treatment, faint bands can be detected in gun1 but not in the wild type. Actin and the chloroplast chaperone protein cpHSC70 were included as loading controls.