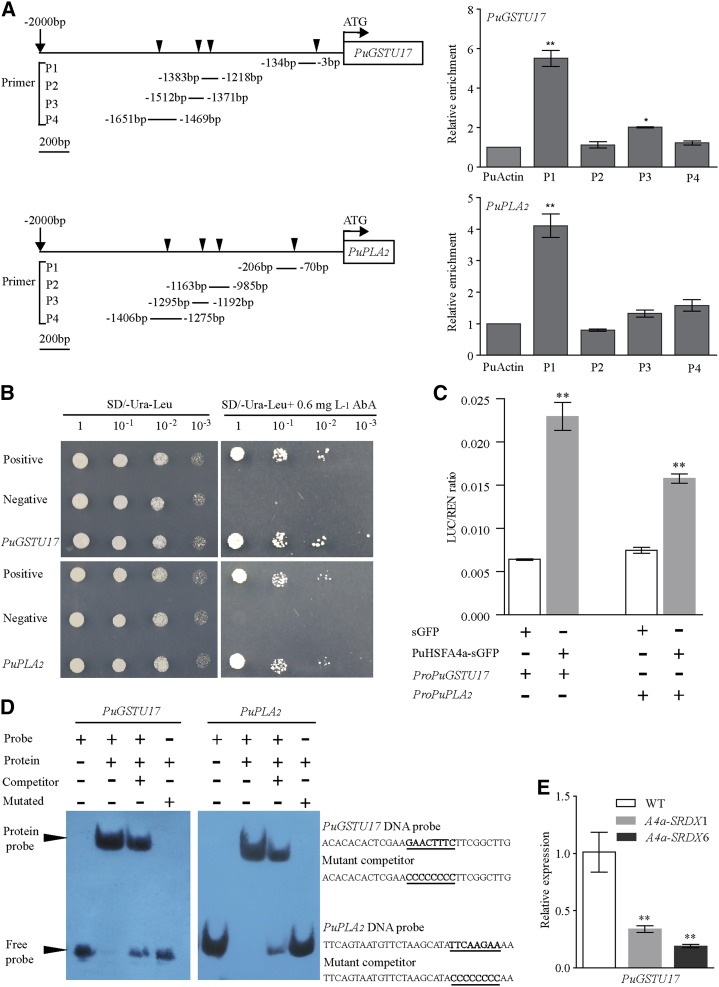
Figure 7.
Verification of the PuHSFA4a target genes PuGSTU17 and PuPLA2. A, ChIP-qPCR analysis of PuHSFA4a binding to PuGSTU17 and PuPLA2 promoter fragments using anti-GFP tag antibody. Schematic diagrams showing the PuHSFA4a binding sites in the regions 2,000 bp upstream of the transcriptional start (ATG) sites of the PuGSTU17 and PuPLA2 genes. Triangular arrowheads indicate PuHSFA4a binding sites (P1, P2, P3, and P4). B, Y1H assay of PuHSFA4a binding to PuGSTU17 and PuPLA2 promoter fragments. C, Binding of PuHSFA4a to the promoters of PuGSTU17 and PuPLA2 as assayed using the dual luciferase system. D, EMSA analyses showing the binding of PuHSFA4a to the DNA probes of the PuGSTU17 and PuPLA2 promoters in vitro. The free and bound DNAs (arrows) were separated by acrylamide gel electrophoresis. The unlabeled probes were used as competitors, and mutated probes were produced by replacing the HSE motifs with GGGGGGGG. E, Relative expression levels of PuGSTU17 in PuHSFA4a-SRDX and wild-type plants assessed under excess Zn conditions and normalized to PuActin (MH644084). In (A), (C), and (E), data are presented as means of three biological replicates, and error bars = sd. Asterisks indicate significant differences as determined by a Student’s t test, *P < 0.05 and **P < 0.01.