Fig. 3.
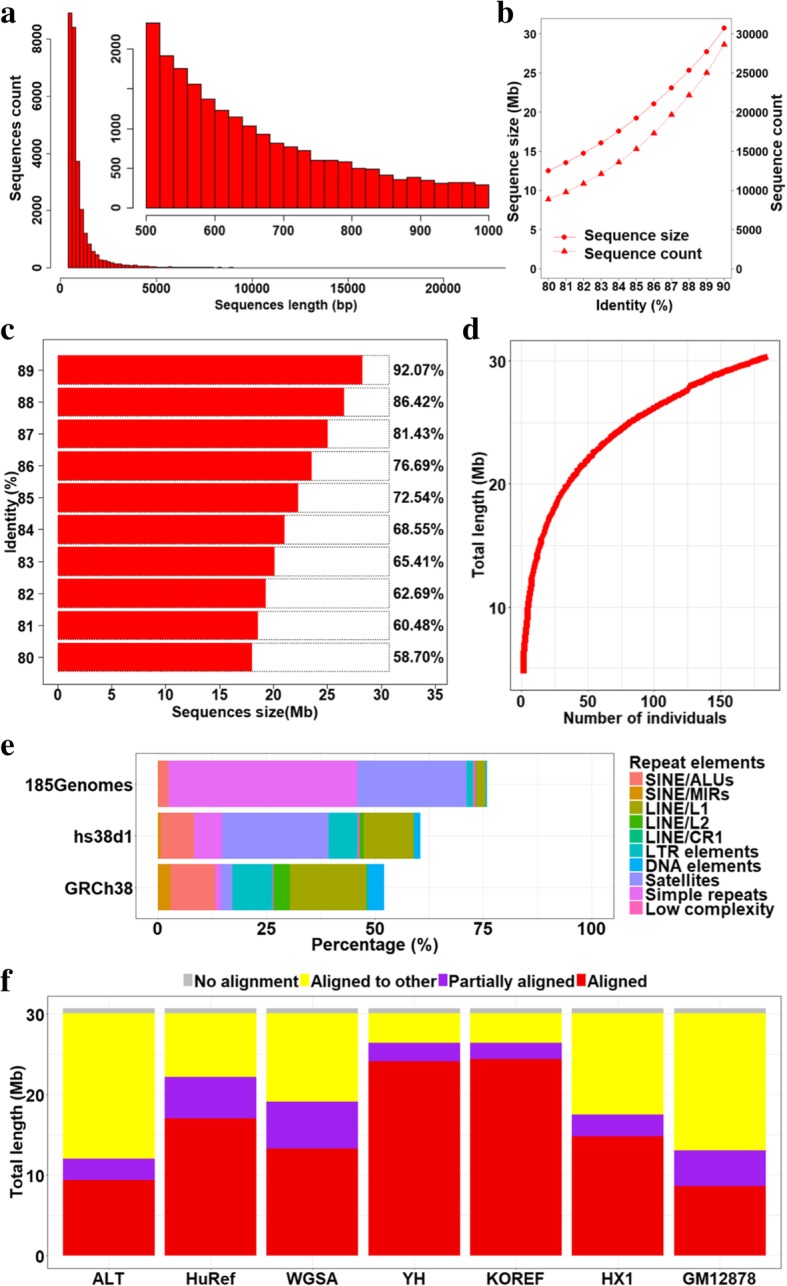
Characterization of sequences fully unaligned to GRCh38 primary assembly sequences in 185 deep sequencing Han Chinese genomes. a Length distribution of fully unaligned sequences. b The total length of fully unaligned sequences (Mb) obtained by using lower identity (80–90%) to remove redundant sequences. c The sequence count and sequence size when aligning the sequences to GRCh38 primary assembly sequences with lower sequence identity (80–90%). d Simulation of the total fully unaligned sequences using different numbers of individuals. e The percentage of repeat elements resulted from RepeatMasker, “hs38d1” is 5.8 Mb novel sequences from SGDP, and “GRCh38” is the primary assembly sequences of the human reference genome GRCh38. The RepeatMasker masked result of GRCh38 was downloaded from http://www.repeatmasker.org/species/hg.html. f Validation of fully unaligned sequences by aligning to other available human sequences (≥ 90% identity). “Aligned” defines the sequences that could be aligned to the target sequences, “Partially aligned” defines the sequences that could be partially aligned to the target sequences, “Aligned to other” defines the sequences that could not be aligned to the target sequences but could be aligned to other six available human sequences, and “No alignment” defines the sequences that could not be aligned to anyone of the seven data sets
