Figure 1.
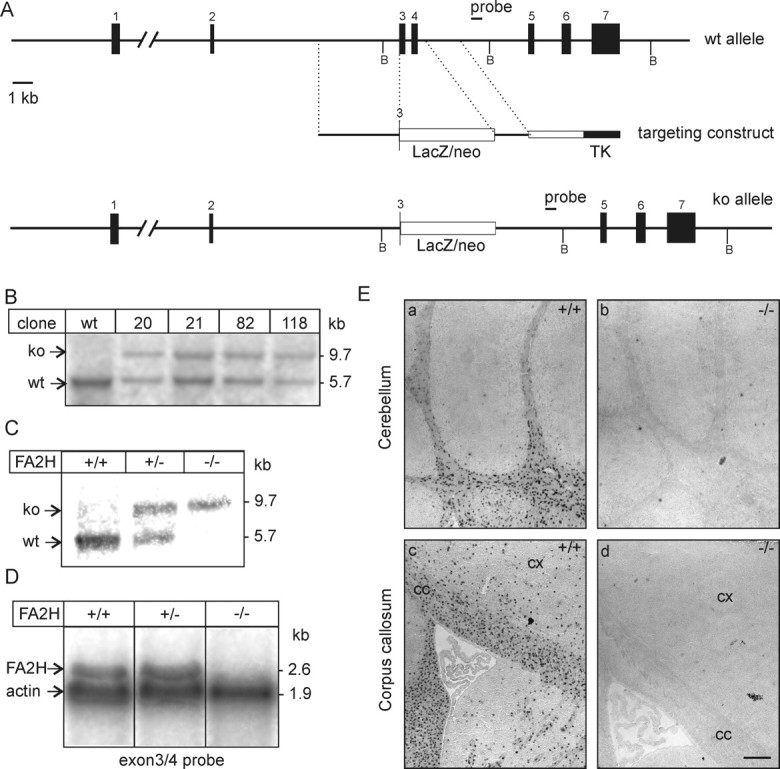
Generation of FA2H-deficient mice. A, Schematic representation of the FA2H wild-type allele, structure of the targeting vector, and targeted allele. Restriction sites for BglII (B) are indicated. B, Southern blot analysis of FA2H+/− ES cell clones, which survived G418/ganciclovir selection. Genomic DNA was digested with BglII and hybridized to the 3′ external probe, indicated in A. A nontargeted ES cell clone (wt) was included as control. ko, Knock-out. C, Southern blot analysis of FA2H alleles from mouse tail genomic DNA digested with BglII. D, Northern blot analysis of FA2H expression. Brain total RNA (20 μg/lane) of 6-week-old FA2H+/+, FA2H+/−, and FA2H−/− mice was separated in 1 m formaldehyde/1% agarose gels and transferred onto nylon membranes. Membranes were simultaneously hybridized to [32P]-labeled FA2H (derived from exons 3 and 4) and β-actin probes. Bound probes were visualized by autoradiography on x-ray films. E, In situ hybridization. Parasagittal sections of 4-week-old mouse brain of FA2H+/+ (a, c) and FA2H−/− (b, d) tissue sections were hybridized to a DIG-labeled FA2H-antisense probe. Shown are the hybridization signals in cerebellum (a, b) and forebrain (c, d). No FA2H mRNA-positive cells were observed in brain sections of FA2H−/− mice. cx, Cortex; cc, corpus callosum. Scale bar, 200 μm.
