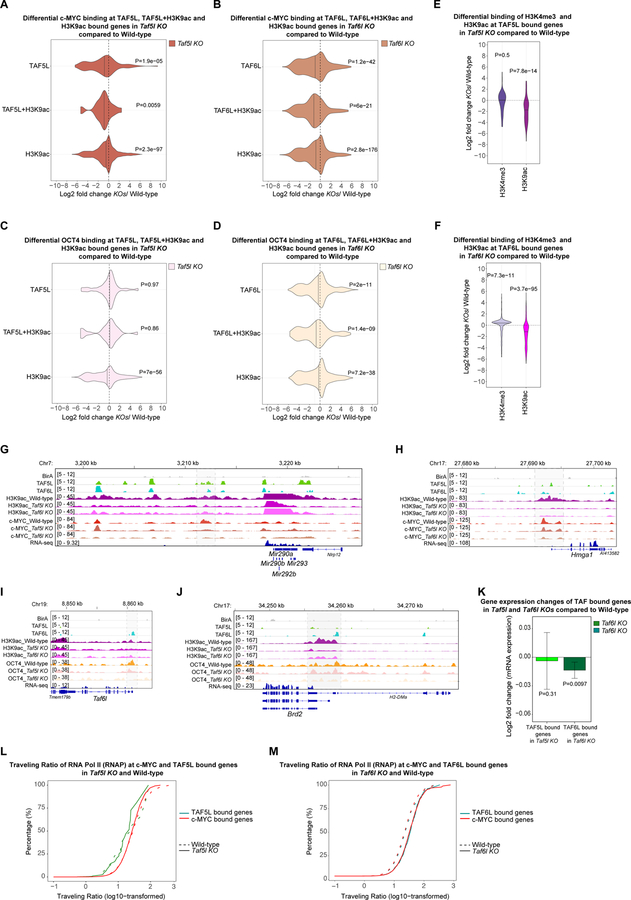
Figure 5. Predominantly TAF5L/TAF6L modulate H3K9ac deposition and c-MYC recruitment at the TAF5L/TAF6L target genes to activate their gene expression through RNA Pol II pause release.
(A) Differential binding of c-MYC at the TAF5L, TAF5L+H3K9ac and H3K9ac bound genes in Taf5l KO compared to wild-type.
(B) Differential binding of c-MYC at the TAF6L, TAF6L+H3K9ac and H3K9ac bound genes in Taf6l KO compared to wild-type.
(C) Differential binding of OCT4 at the TAF5L, TAF5L+H3K9ac and H3K9ac bound genes in Taf5l KO compared to wild-type.
(D) Differential binding of OCT4 at the TAF6L, TAF6L+H3K9ac and H3K9ac bound genes in Taf6l KO compared to wild-type.
(E) Differential binding of H3K9ac and H3K4me3 at the TAF5L bound genes in Taf5l KO compared to wild-type.
(F) Differential binding of H3K9ac and H3K4me3 at the TAF6L bound genes in Taf6l KO compared to wild-type.
(G, H) Genomic tracks of H3K9ac and c-MYC binding at miR 290–295 cluster and Hmga1 gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA (control), TAF5L and TAF6L binding are presented at the same gene loci. Highlighted regions show changes of H3K9ac and c-MYC binding.
(I, J) Genomic tracks of H3K9ac and OCT4 binding at Taf6l and Brd2 gene loci from wild-type, Taf5l and Taf6l KOs. Genomic tracks of BirA (control), TAF5L and TAF6L binding are presented at the same gene loci. Highlighted regions show changes of H3K9ac and OCT4 binding.
(K) Gene expression changes of TAF5L and TAF6L bound genes in Taf5l and Taf6l KOs, respectively.
(L) The traveling ratio (TR) of RNA Pol II (RNAP) at c-MYC and TAF5L bound genes in Taf5l KO and wild-type.
(M) The traveling ratio (TR) of RNA Pol II (RNAP) at c-MYC and TAF6L bound genes in Taf6l KO and wild-type.
Related to Figure S5.