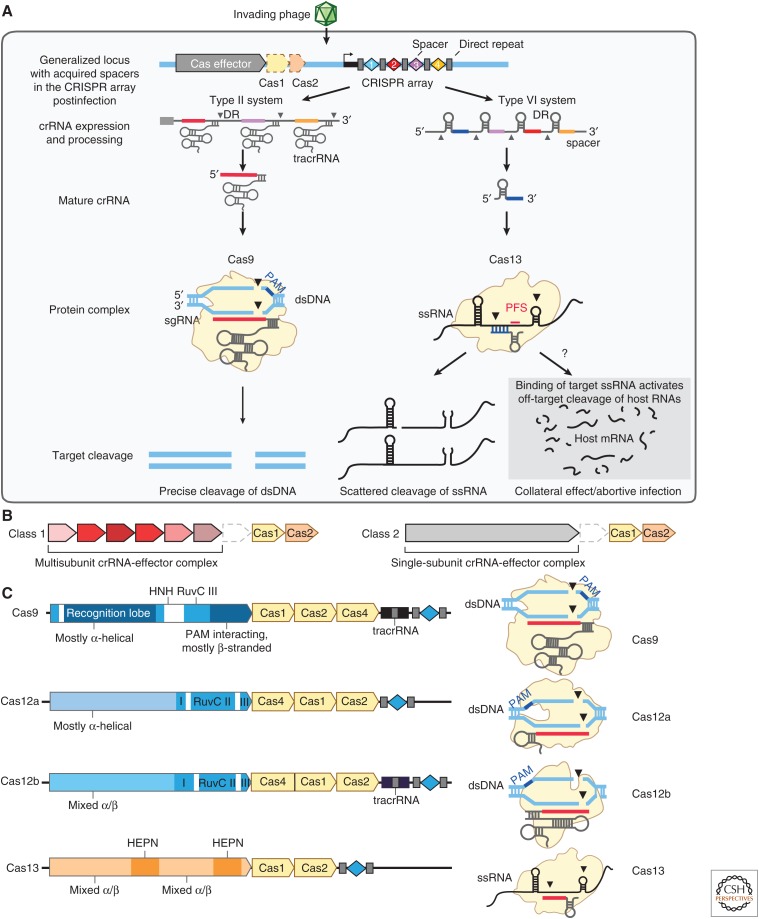
Figure 2.
Structure and function of CRISPR systems. (A) Depiction of the three steps of CRISPR immunity (adaptation, expression, and interference) by DNA-targeting (e.g., Cas9-containing type II) and RNA-targeting (e.g., Cas13-containing type VI) systems. Cas9 interference is mediated by RNA-guided cleavage of double-stranded DNA (dsDNA) targets. Cas13 interference results in two modes of activation in which RNA-guided targeting of single-stranded RNAs (ssRNAs) results in targeted cleavage of the transcripts as well as collateral cleavage of noncomplementary RNAs that are in proximity to the activated Cas13 complex. The collateral cleavage of Cas13 systems is thought to be part of a programmed cell death pathway to further augment immunity against pathogen infection. (B) CRISPR systems can be divided into two classes: Class 1 systems consist of multiprotein interference complexes (types I, III, and IV systems), whereas Class 2 systems contain a single-protein interference effector (types II, V, and VI systems). (C) Genetic loci for four Class 2 CRISPR systems (left), including sequences coding for effector proteins annotated with key protein domains, accessory Cas proteins involved in adaptation colored in beige, trans-activating CRISPR RNAs (tracrRNAs) (if required), and CRISPR array. The protein complexes (right) show the nucleic acid target (dsDNA or ssRNA), locations of the protospacer adjacent motif (PAM) sequence and cleavage sites, and approximate structure of the guide RNA.