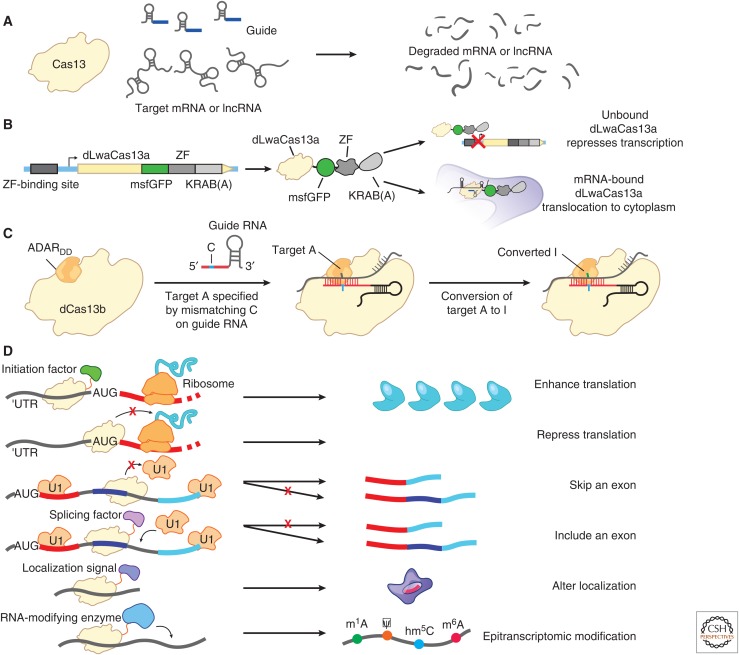
Figure 7.
RNA targeting applications of Cas13. (A) RNA knockdown with Cas13. (B) Image tracking using dCas13-eGFP to fluorescently visualize transcripts in cells. Modifying the dCas13-EGFP fusion with a zinc finger–Krüppel associated box (KRAB) fusion can improve visualization and lower fluorescence background signal by repressing the expression of unbound dCas13-EGFP complexes via negative feedback. (C) RNA editing of adenines to inosines by a dCas13-ADAR2 fusion. Precise editing is accomplished by creating an A:C base pair bubble by encoding a mismatch in the guide RNA. (D) Further applications of Cas13 for studying RNA regulation. Translation could be activated by recruiting translation initiation factors or repressed by blocking start codons on transcripts. Splicing has been altered by blocking splice sites on transcripts or by recruiting splicing factors. Transcript localization could be modulated by fusing dCas13 to different localization sequences. Lastly, posttranscriptional RNA modifications could be introduced by recruiting RNA-modifying enzymes, such as pseudouridine synthase, to specific bases on RNA transcripts.