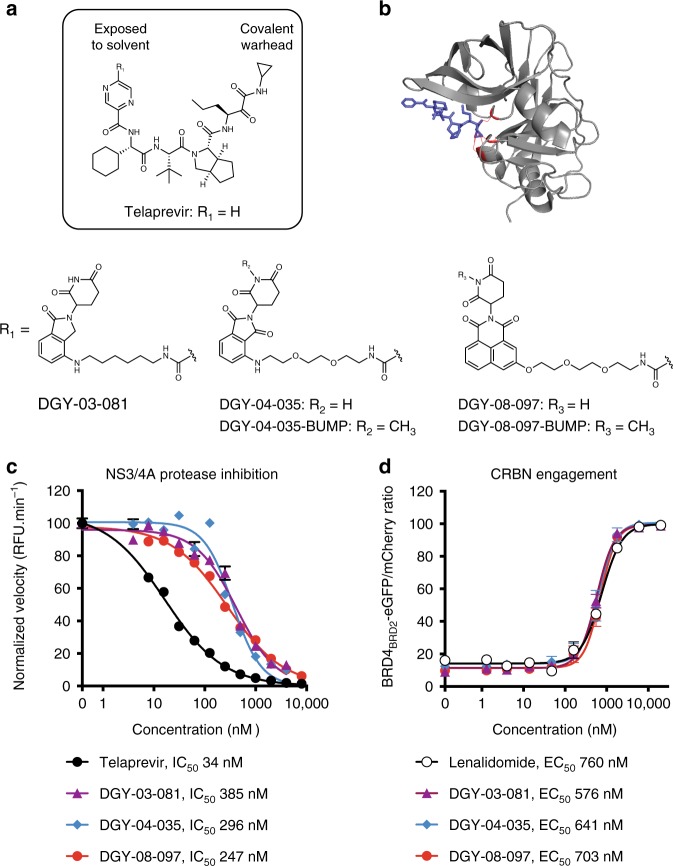
Fig. 1.
Design and biochemical characterization of the NS3-targeting degraders a Chemical structures of telaprevir and the degrader derivatives DGY-03-081, DGY-04-035, and DGY-08-097 where R1 is the CRBN ligand. DGY-04-035-BUMP and DGY-08-097-BUMP are negative control analogs due to methylation of the glutarimide, as this modification has previously been shown to ablate CRBN-binding11. b Crystal structure of telaprevir bound to the HCV NS3/4A protease complex (PDB 3SV6). Telaprevir is shown in blue and the protease catalytic triad is shown in red. c Measurement of NS3/4A protease inhibition in vitro. Cell lysates containing the endogenous NS3/4A protease were prepared from cells stably expressing an HCV subgenomic replicon. These were incubated with candidate degraders at the indicated concentrations followed by addition of an HCV NS3 FRET peptide substrate. Cleavage of the FRET substrate was measured for 1 h at 30 °C, and the rate of enzymatic cleavage was fitted by linear regression. The concentration that led to a 50% decrease in HCV NS3/4A enzymatic activity (IC50) was determined by nonlinear regression. Data are presented as means normalized to DMSO ± standard deviation of n = 2 technical replicates. One representative experiment is shown, with IC50 values averaged from n ≥ 3 independent experiments. Source data are provided as a Source Data file. RFU: relative fluorescence units. d Quantitative assessment of intracellular CRBN engagement using a BRD4BRD2-eGFP-mCherry reporter assay26. CRBN engagement was assessed by monitoring rescue of dBET6-mediated degradation of BRD4BRD2-eGFP. Cells stably expressing BRD4BRD2-eGFP and mCherry were treated with 100 nM of dBET6, a specific degrader of BRD4BRD2, and increasing concentrations of the candidate NS3 degrader. The eGFP and mCherry signals were quantified by flow cytometry analysis, and the concentration of compound that rescued 50% of BRD4BRD2-eGFP fluorescence (EC50) was determined by nonlinear regression. Data are presented as means normalized to DMSO ± standard deviation of n = 3 technical replicates. One representative experiment of n ≥ 2 is shown