Figure 1.
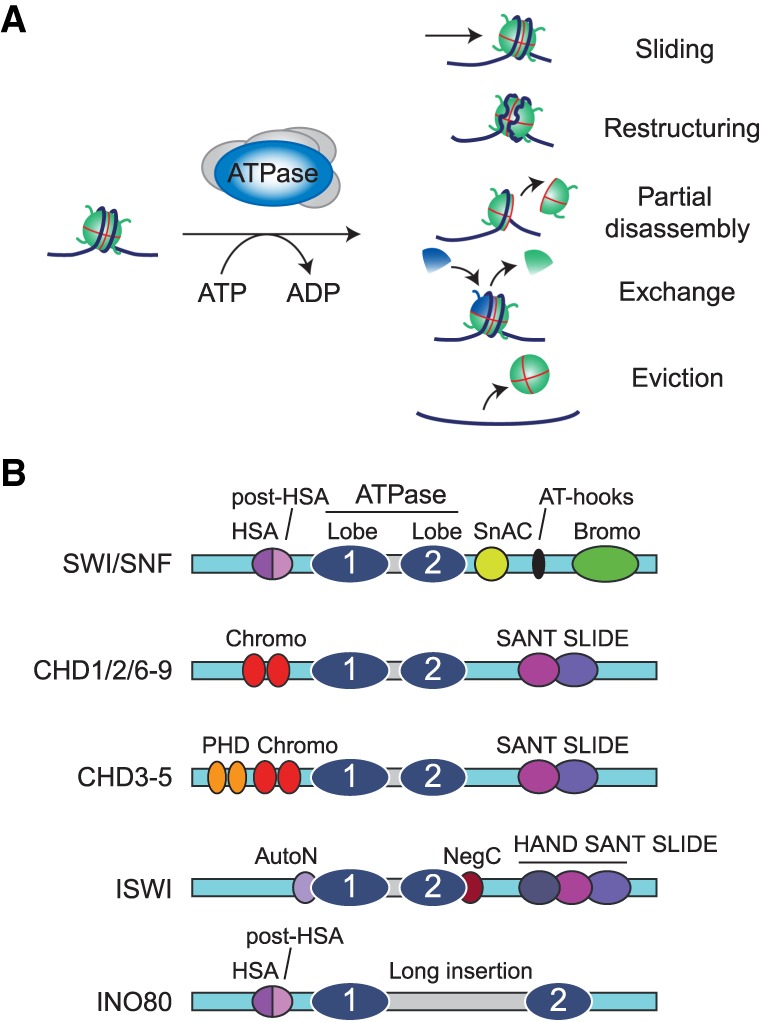
ATP-dependent chromatin remodeling. (A) Different outcomes of ATP-dependent remodeling of nucleosomes. Remodeler action can drive the sliding of a nucleosome to another position on the DNA, thus exposing a previously bound sequence. Alternatively, remodelers can make the nucleosomal DNA more accessible, while the histone octamer remains associated. Remodeling can also disrupt the octamer structure causing a partial disassembly, typically through eviction of histone H2A/H2B dimers. Specialized remodelers can mediate the exchange between histone variants. Finally, remodeling can result in the complete eviction of the histone octamer. (B) Structural domains of the four major Snf2 ATPase subfamilies, SWI/SNF, CHD, ISWI, and INO80. The translocase/ATPase domain of all remodelers comprises two RecA-like lobes separated by an insertion (highlighted in gray). Members of the INO80 family have a longer insertion than other remodelers. Each subfamily is characterized by a unique set of additional domains, including the HSA (helicase SANT-associated) and post-HSA domains, SnAC (Snf2 ATP coupling), AT hooks (A/T-rich DNA-binding domains), Bromo (bromodomains), Chromo (chromodomains), SANT-SLIDE domain, PHD finger (plant homeodomain), HAND-SANT-SLIDE domain, AutoN (autoinhibitory N-terminal), and NegC (negative regulator of coupling). See the text for details and references.
