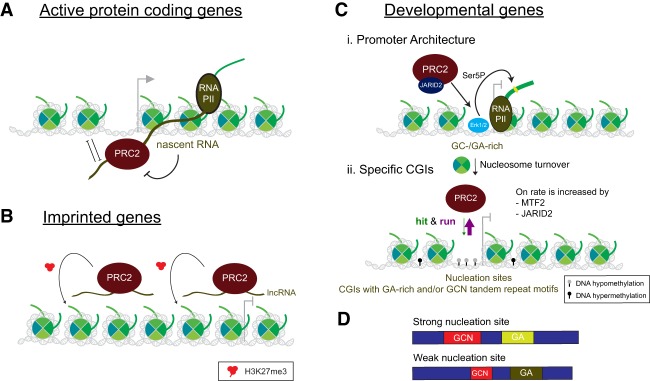
Figure 7.
Three modes of PRC2 recruitment to chromatin in mammals. (A) At active protein coding genes, the 5′ region of the nascent RNA recruits PRC2 through interaction with its core EZH2 subunit. This interaction precludes PRC2 activity. (B) At imprinted genes, long ncRNAs (lncRNAs) produced from the same loci recruit PRC2 in cis. PRC2 deposits H3K27me3 on these transcriptionally silent genes. (C) At developmental genes, promoter architecture and specific CGIs recruit PRC2. (Panel i) Erk1/2 localizes to GC-/GA-rich regions on the genome, mediating nucleosome turnover as well as phosphorylation of RNAPII at its C-terminal domain (CTD)-Ser5 residue. These events provide a promoter architecture conducive to PRC2/JARID2 recruitment. (Panel ii) Through its low-affinity interactions with chromatin, PRC2 can recognize nucleation sites that contain hypomethylated CGIs with GA-rich and/or GCN tandem repeat motifs via a “hit and run” mechanism, but its on rate is lower than its off rate. The on rate is increased by PRC2 interaction with MTF2/PCL2 and/or JARID2. (D) Nucleation sites (both strong and weak) are enriched for dense CGIs, which have a high number of CG dinucleotides within the island (blue boxes). Both types of nucleation sites are enriched for “GA” and/or “GCN” tandem repeat motifs; however, strong nucleation sites have longer GCN tandem repeats (red boxes) and a different distribution of GA content (light-green and dark-green boxes) compared with weak ones.