Figure 1.
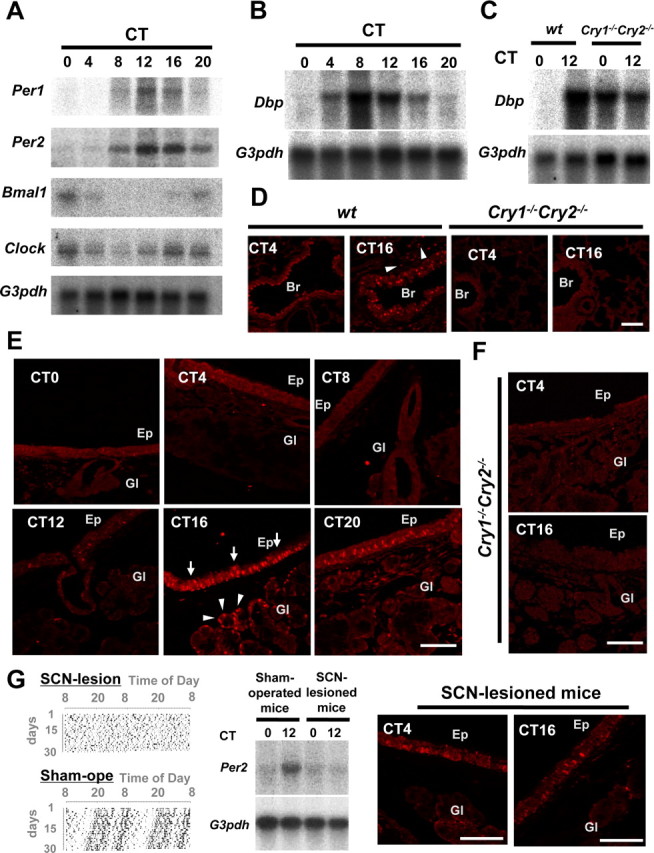
Clock genes and clock controlled genes in the mouse respiratory system. A, Circadian expression of Per1, Per2, Bmal1, and Clock genes, as detected by Northern blot analysis. G3pdh expression was determined as a control. Note the peak and through in Per1 and Per2 mRNA levels at CT12 and CT0, respectively, as well as the inverted rhythms of Bmal1 and Clock gene expression. B, Circadian expression of Dbp, a clock controlled gene. Dbp mRNA levels show a similar distribution pattern as that of Per1. C, Dbp mRNA levels in arrhythmic Cry1−/−Cry2−/− mice. D, Immunohistochemical staining of the PER2 protein in the lung and bronchiole of wild-type and Cry1−/−Cry2−/− mice at CT4 and CT16. PER2 proteins were abundantly expressed in the nucleus of bronchial epithelial cells at CT16, although almost absent at CT4. Alveolar cells (arrowheads) also showed a slight rhythm. PER2 protein expression was abolished in Cry1−/−Cry2−/− mice at any time examined. E, Circadian expression of the PER2 protein in epithelium and submucosal glands in trachea. Note the high level of nuclear immunohistochemical staining in tracheal epithelium (arrows) and submucosal glands (arrowheads) at CT16. F, Abolished PER2 protein expression in Cry1−/−Cry2−/− mice. G, Loss of circadian oscillation in Per2 mRNA and PER2 protein levels in the respiratory system of SCN-lesioned animals. Shown are (double-plotted) behavioral actograms (left), and Per2 mRNA expression levels at CT0 and CT12, as determined by Northern blot analysis (middle) of SCN-lesioned and sham-operated wild-type mice, as well as PER2-immunohistochemistry in SCN-lesioned mice at CT4 and CT16. Data were adopted as control versus sham-operated animals. Br, Bronchiole; Ep, tracheal epithelium; Gl, submucosal gland. Scale bars, 100 μm.
