Figure 5.
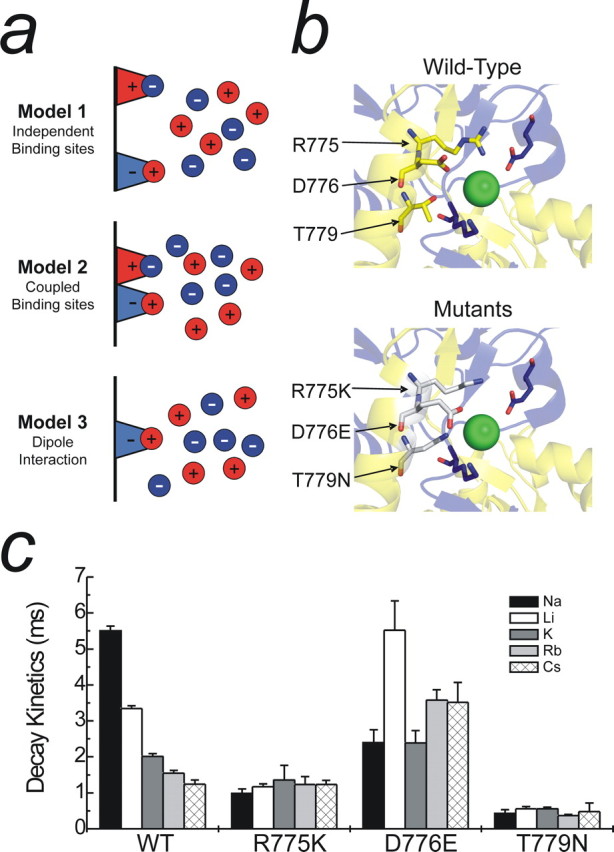
Possible mechanisms for monovalent ion interactions at KARs. a, Schematic showing three distinct models to explain the effect of monovalent anions and cations on GluR6 KARs. b, Top, Crystal structure showing critical amino acid residues that constitute the proposed anion binding site (Protein Data Bank number 2F34). Although only one dimer is shown, the ion is also conjugated by the corresponding amino acids from the adjacent subunit. b, Bottom, Proposed anion binding site containing point mutations (R775K, D776E, and T779N) that interfere with both anion and cation modulation of GluR KARs. Note that each mutation elicits an enlargement of the anion binding pocket through changes in the orientation of each residue. c, The effect of changing external cation species with the various anion binding site mutants. Both R775K and T779N abolish cation modulation, whereas cation modulation is markedly reduced in the D776E mutant. Data are mean ± SEM of at least three patches per mutant in each cation.
