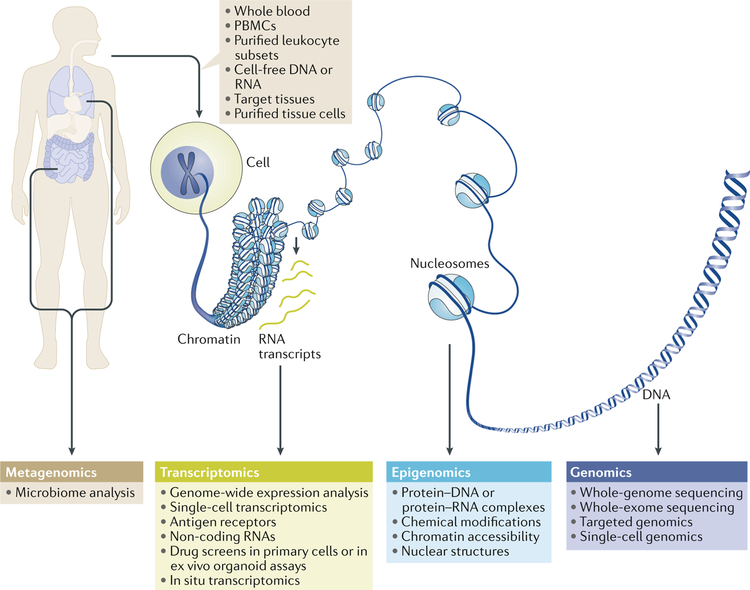
Fig. 1 |. Overview of next-generation sequencing applications.
Large amounts of transcriptomic, epigenomic, genomic and metagenomic sequencing data can be generated from an individual. Sources of nucleic acids for sequencing include cells obtained from blood or tissues and cell-free DNA or RNA. Microbiome analysis (metagenomics) can be performed on microorganisms from various tissues including the gut, skin and urogenital tract. Linking strong hypotheses with the most informative and robust techniques in carefully characterized cohorts of patients holds promise for potentially groundbreaking discoveries in rheumatic diseases. PBMCs, peripheral blood mononuclear cells.