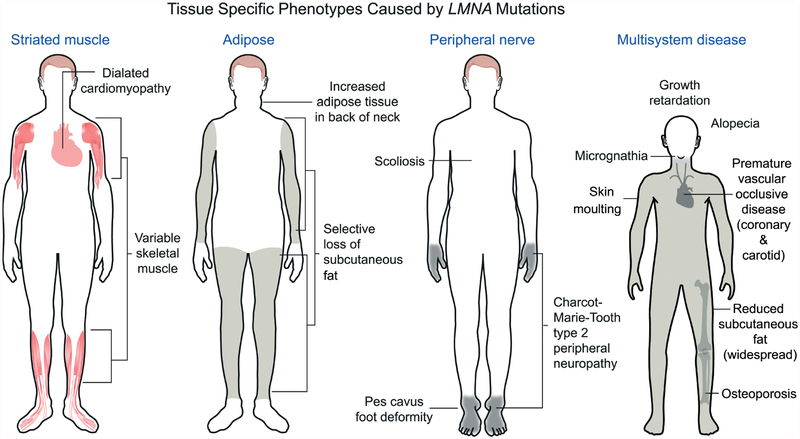
Figure 2.
Different LMNA mutations cause diseases that affect striated muscle, adipose, peripheral nerve or multiple systems with features of accelerated ageing. Most LMNA mutations are autosomal-dominant and cause dilated cardiomyopathy with variable skeletal muscle involvement. This includes the classical Emery–Dreifuss muscular dystrophy phenotype, as shown in the diagram, with scapulohumeral–peroneal distribution of skeletal muscle involvement, concurrent tendon contractures and dilated cardiomyopathy. Autosomal-dominant missense mutations in LMNA, the large majority of which cause a change in the surface charge of the immunoglobulin-like fold of lamin A and lamin C, cause Dunnigan-type familial partial lipodystrophy, with selective loss of subcutaneous fat from the extremities, fat accumulation in the neck and face and insulin resistance and diabetes mellitus. An autosomal recessive LMNA mutation that leads to an arginine-to-cysteine amino acid substitution at residue 298 causes a Charcot–Marie–Tooth type 2 peripheral neuropathy, characterized clinically by a stocking–glove sensory neuropathy, resultant pes cavus foot deformity and other variable features, such as scoliosis. The sporadic cytosine to thymine transversion in codon 608 of exon 11 of LMNA causes Hutchinson–Gilford progeria syndrome, which has features of accelerated ageing, such as sclerotic skin, joint contractures, micrognathia, alopecia, fingertip tufting, distal-joint abnormalities, growth impairment and vascular abnormalities, generally leading to death during the second decade due to myocardial infarction or stroke. Other LMNA mutations can also cause progeroid syndromes with similar features; a recessive LMNA mutation causing an arginine-to-histidine amino acid substitution at residue 527 in the immunoglobulin-like fold causes mandibuloacral dysplasia, a disorder with a combination of progeroid features and partial lipodystrophy. Reproduced with permission from: Dauer WT, Worman HJ. The nuclear envelope as a signalling node in development and disease. Dev Cell 2009; 17: 626–638 [65].