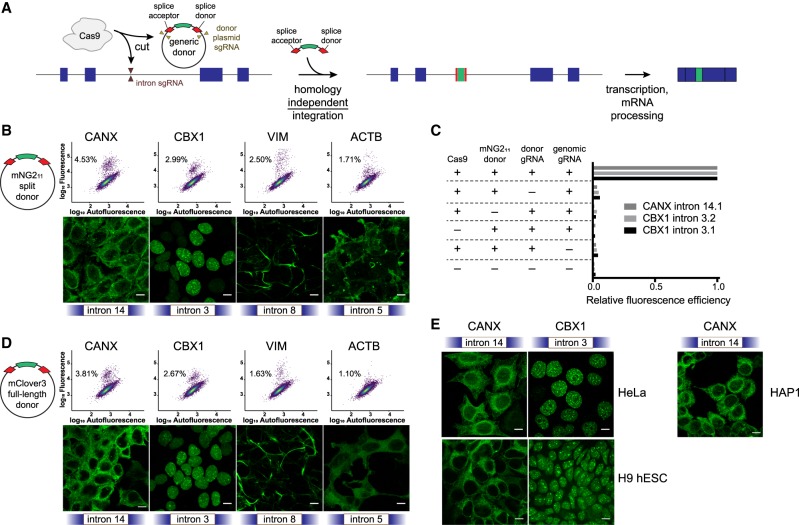
Figure 1.
Homology-independent generic intron tagging enables efficient and easy generation of endogenous fusions. (A) Illustration of the tagging approach: Double-strand breaks are generated in the intron and donor resulting in the addition of a synthetic exon and fusion of the tag to the coding sequence. (B) Using a small donor composed of the mNG211 epitope flanked by splice acceptor and donor sites results in efficient tagging of CANX, CBX1, VIM, and ACTB at the indicated introns (all sgRNA “target 1”), as observed by flow cytometry (upper panels, colored by density) and by confocal microscopy (lower panels). Percentages in the dot plots represent the green population as a subset of the total. (C) All transfection mix components are required for tagging of CANX intron 14, sgRNA target 1 (14.1), and of CBX1 intron 3, targets 1 and 2 (3.1 and 3.2). The table indicates which component was removed, and bar plots represent the relative percentage of fluorescence-positive cells compared to the full mix. (D) Tagging using a full-length mClover3 fluorophore as a donor. (E) Tagging of CANX and CBX1 in HeLa cells, H9 human embryonic stem cells (hESC), and HAP1 cells. All images are maximum projections of Z-stacks, and scale bars correspond to 10 µm.