Figure 2.
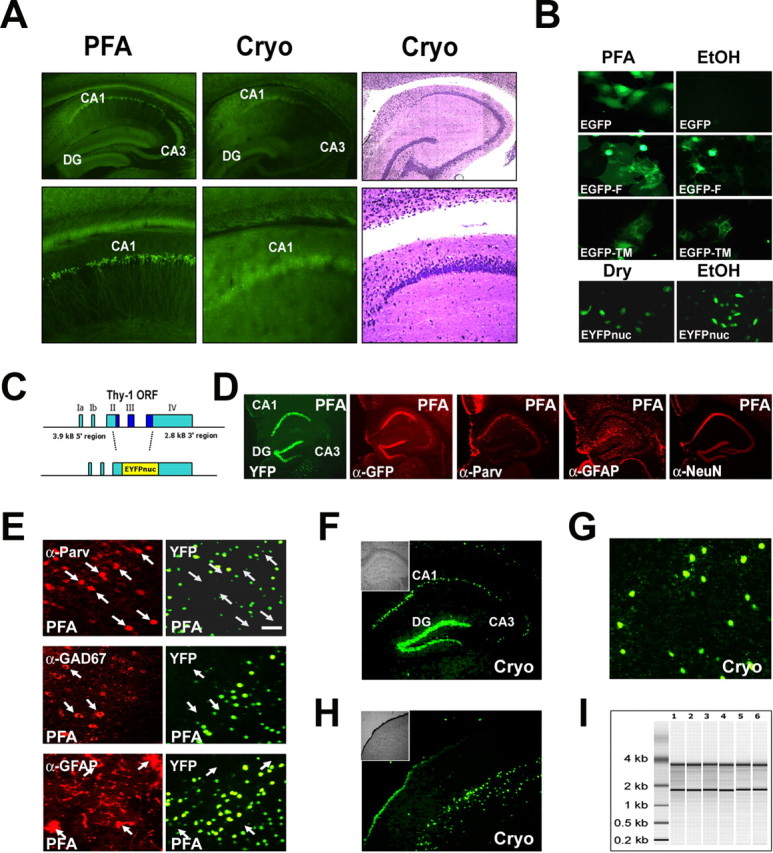
Conventional GFP derivatives are not compatible with laser-directed microdissection. The cellular signal of a nuclear-targeted EYFP (EYFPnuc) is preserved in freeze-dried cryosections obtained from transgenic mice. RNA isolated from cryosections is intact. A, The cellular resolution of EYFP expressed in transgenic mice is lost after cryosectioning. Sagittal sections of mouse brains from mice expressing cytoplasmic localized EYFP under the control of the Thy-1 promoter were either fixed with 4% buffered PFA or immediately frozen on dry ice. Vibratome sections (PFA, left) obtained from PFA fixed tissue (30 μm) and cryosections (15 μm; Cryo, middle) from frozen tissue, were analyzed for EYFP fluorescence. After cryosectioning, the cellular resolution of the EYFP fluorescent signal is lost. The EYFP signal is retained but appears to be diffuse and faded. Staining of the cryosection with thionin (Cryo, right) reveals that the cellular integrity of the tissue specimen is intact. B, Nuclear targeted fluorescent proteins appear to be promising candidates for a combined in vivo labeling and microdissection approach. COS7 or HeLa cells were transiently transfected with expression constructs coding for GFP derivatives localized to different cellular compartments: EGFP (cytoplasmic localization), EGFP carrying a farnesylation signal (EGFP-F, attached to membranes), EGFP fused to the N terminus of a synthetic transmembrane protein (TM-EGFP), and EYFP fused to three nuclear localization signals (EYFPnuc). Forty-eight hours after transfection, COS7 cells were either fixed with a PBS-buffered 4% PFA or fixed with 70% ethanol (EtOH) and subsequently dry mounted. HeLa cells transfected with EYFPnuc were either dry mounted (DRY) or fixed with 70% ethanol (EtOH) and subsequently frozen at −80°C before analysis. C, Transgenic mice were generated that express a nuclear-targeted EYFP (EYFPnuc) under control of the Thy-1 Promoter (TYNC mice). The Thy-1 minigene was modified by replacing the Thy-1 ORF with sequences coding for EYFPnuc. D, E, Vibratome sections (30 μm) from brains of PFA-fixed TYNC mice were analyzed for YFP fluorescence and for cellular marker gene expression with immunohistochemistry. D, Within the hippocampus, the YFP expression is most prominent in pyramidal neurons of the CA1 field (CA1) and in the granular cell layer of the dentate gyrus (DG). In the CA3 field (CA3), fewer cells are YFP positive. The YFP fluorescence appears to be strictly overlapping with the indirect GFP immune fluorescence analysis (α-GFP), although it is less sensitive. The pan-neuronal marker NeuN (α-NeuN) colocalizes with the nuclear YFP signal. Staining with GFAP (α-GFAP) and parvalbumin (α-Parv) does not reveal YFP overlapping signals. E, Higher-magnification photographs obtained from vibratome sections at the level of the primary motor and sensory cortex. YFP-positive nuclei (YFP) do not colocalize with the interneuronal marker parvalbumin (α-Parv) and GAD67 (α-GAD67) and also not with the astrocyte marker GFAP (α-GFAP). Arrows mark parvalbumin, GAD67, or GFAP-positive cells stained in red in the left panel. No overlap with YFP fluorescence can be detected in the right panel with arrows pointing to identical positions as in the left panel. Scale bar, 50 μm. F–H, Cryosections (8 μm; Cryo) were mounted on PEN foils (Leica) and analyzed for YFP fluorescence. F, In the hippocampus, YFP-positive nuclei can be detected in the granular cell layer (DG) and in the CA1 field of the pyramidal cell layer (CA1). In the CA3 field (CA3), fewer cells are YFP positive. The inset shows the morphology of the hippocampus in bright field. G, Higher magnification of YFP-positive nuclei in the somatosensory cortex. Scale bar, 50 μm. H, In the cortex, cells located in deeper layers show a nuclear YFP signal. The inset shows the morphology of the section at the level of the somatosensory cortex in bright field. I, Total RNA analyzed with a Bioanalyzer (Agilent); each lane represents half of the RNA isolated from one coronal. brain section. Adjacent 8-μ-thick sections were cryomounted on PEN foil slides, dried, and treated as follows: (1) stored for 2 h at room temperature, (2) stored for 8 h at room temperature, (3) stored for 24 h at room temperature, (4) stored for 48 h at room temperature, (5) frozen at −80°C and then kept at room temperature for 2 h, and (6) microdissected regions pooled from eight sections (total area size isolated comparable with one section). The ratio of the 28S versus the 18S rRNA bands was determined with the Bioanalyzer software and for all samples was 1.3± 0.1. The amount of RNA isolated from one coronal 8 μ brain cryosection was ∼50 ng.
